Exploring Cuba's population structure and demographic history using genome-wide data
- PMID: 30061702
- PMCID: PMC6065444
- DOI: 10.1038/s41598-018-29851-3
Exploring Cuba's population structure and demographic history using genome-wide data
Abstract
Cuba is the most populated country in the Caribbean and has a rich and heterogeneous genetic heritage. Here, we take advantage of dense genomic data from 860 Cuban individuals to reconstruct the genetic structure and ancestral origins of this population. We found distinct admixture patterns between and within the Cuban provinces. Eastern provinces have higher African and Native American ancestry contributions (average 26% and 10%, respectively) than the rest of the Cuban provinces (average 17% and 5%, respectively). Furthermore, in the Eastern Cuban region, we identified more intense sex-specific admixture patterns, strongly biased towards European male and African/Native American female ancestries. Our subcontinental ancestry analyses in Cuba highlight the Iberian population as the best proxy European source population, South American and Mesoamerican populations as the closest Native American ancestral component, and populations from West Central and Central Africa as the best proxy sources of the African ancestral component. Finally, we found complex admixture processes involving two migration pulses from both Native American and African sources. Most of the inferred Native American admixture events happened early during the Cuban colonial period, whereas the African admixture took place during the slave trade and more recently as a probable result of large-scale migrations from Haiti.
Conflict of interest statement
The authors declare no competing interests.
Figures
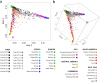

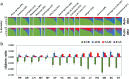


References
-
- Boomert, A. The Caribbean Islands. In The Cambridge World Prehistory (eds Renfrew, C. & Bahn, P.) 2, 1217–1234 (Cambridge University Press, 2014).
-
- Fitzpatrick SM. The Pre-Columbian Caribbean: Colonization, Population Dispersal, and Island Adaptations. PaleoAmerica. 2015;1:305–331. doi: 10.1179/2055557115Y.0000000010. - DOI
-
- Granberry, J. & Vescelius, G. Languages of the Pre-Columbian Antilles. (University of Alabama Press, 2004).
-
- Perez de la Riva, J. P. A World Destroyed. In TheCuba Reader (eds Chomsky, A., Carr, B., Smorkaloff, P. M., Kirk, R. & Starn, O.) 20–25 (Duke University Press, 2003). 10.1215/9780822384915-005.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

