Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis
- PMID: 30061742
- PMCID: PMC6123400
- DOI: 10.1038/s41422-018-0074-y
Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis
Abstract
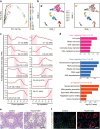
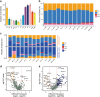
A systematic interrogation of male germ cells is key to complete understanding of molecular mechanisms governing spermatogenesis and the development of new strategies for infertility therapies and male contraception. Here we develop an approach to purify all types of homogeneous spermatogenic cells by combining transgenic labeling and synchronization of the cycle of the seminiferous epithelium, and subsequent single-cell RNA-sequencing. We reveal extensive and previously uncharacterized dynamic processes and molecular signatures in gene expression, as well as specific patterns of alternative splicing, and novel regulators for specific stages of male germ cell development. Our transcriptomics analyses led us to discover discriminative markers for isolating round spermatids at specific stages, and different embryo developmental potentials between early and late stage spermatids, providing evidence that maturation of round spermatids impacts on embryo development. This work provides valuable insights into mammalian spermatogenesis, and a comprehensive resource for future studies towards the complete elucidation of gametogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures





Comment in
-
Staged profiling of sperm development in sync.Cell Res. 2018 Oct;28(10):965-966. doi: 10.1038/s41422-018-0088-5. Cell Res. 2018. PMID: 30218061 Free PMC article. No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

