Quorum-Quenching Bacteria Isolated From Red Sea Sediments Reduce Biofilm Formation by Pseudomonas aeruginosa
- PMID: 30065702
- PMCID: PMC6057113
- DOI: 10.3389/fmicb.2018.01354
Quorum-Quenching Bacteria Isolated From Red Sea Sediments Reduce Biofilm Formation by Pseudomonas aeruginosa
Abstract
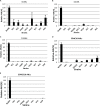
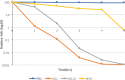
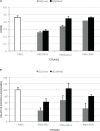
Quorum sensing (QS) is the process by which bacteria communicate with each other through small signaling molecules such as N-acylhomoserine lactones (AHLs). Certain bacteria can degrade AHL molecules by a process called quorum quenching (QQ); therefore, QQ can be used to control bacterial infections and biofilm formation. In this study, we aimed to identify new species of bacteria with QQ activity. Red Sea sediments were collected either from the close vicinity of seagrass or from areas with no vegetation. We isolated 72 bacterial strains, which were tested for their ability to degrade/inactivate AHL molecules. Chromobacterium violaceum CV026-based bioassay was used for the initial screening of isolates with QQ activity. QQ activity was further quantified using high-performance liquid chromatography-tandem mass spectrometry. We found that these isolates could degrade AHL molecules of different acyl chain lengths as well as modifications. 16S-rRNA sequencing of positive QQ isolates showed that they belonged to three different genera. Specifically, two isolates belonged to the genus Erythrobacter; four, Labrenzia; and one, Bacterioplanes. The genome of one representative isolate from each genus was sequenced, and potential QQ enzymes, namely, lactonases and acylases, were identified. The ability of these isolates to degrade the 3OXOC12-AHLs produced by Pseudomonas aeruginosa PAO1 and hence inhibit biofilm formation was investigated. Our results showed that the isolate VG12 (genus Labrenzia) is better than other isolates at controlling biofilm formation by PAO1 and degradation of different AHL molecules. Time-course experiments to study AHL degradation showed that VG1 (genus Erythrobacter) could degrade AHLs faster than other isolates. Thus, QQ bacteria or enzymes can be used in combination with an antibacterial to overcome antibiotic resistance.
Keywords: N-acylhomoserine lactone degradation; Red Sea sediments; biofilm inhibition; marine bacteria; quorum quenching.
Figures

References
-
- Biebl H., Pukall R., Lunsdorf H., Schulz S., Allgaier M., Tindall B. J., et al. (2007). Description of Labrenzia alexandrii gen. nov., sp. nov., a novel alphaproteobacterium containing bacteriochlorophyll a, and a proposal for reclassification of Stappia aggregata as Labrenzia aggregata comb. nov., of Stappia marina as Labrenzia marina comb. nov. and of Stappia alba as Labrenzia alba comb. nov., and emended descriptions of the genera Pannonibacter, Stappia and Roseibium, and of the species Roseibium denhamense and Roseibium hamelinense. Int. J. Syst. Evol. Microbiol. 571095–1107. 10.1099/ijs.0.64821-0 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

