NMR Metabolomics Defining Genetic Variation in Pea Seed Metabolites
- PMID: 30065739
- PMCID: PMC6056766
- DOI: 10.3389/fpls.2018.01022
NMR Metabolomics Defining Genetic Variation in Pea Seed Metabolites
Abstract
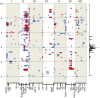
Nuclear magnetic resonance (NMR) spectroscopy profiling was used to provide an unbiased assessment of changes to the metabolite composition of seeds and to define genetic variation for a range of pea seed metabolites. Mature seeds from recombinant inbred lines, derived from three mapping populations for which there is substantial genetic marker linkage information, were grown in two environments/years and analyzed by non-targeted NMR. Adaptive binning of the NMR metabolite data, followed by analysis of quantitative variation among lines for individual bins, identified the main genomic regions determining this metabolic variability and the variability for selected compounds was investigated. Analysis by t-tests identified a set of bins with highly significant associations to genetic map regions, based on probability (p) values that were appreciably lower than those determined for randomized data. The correlation between bins showing high mean absolute deviation and those showing low p-values for marker association provided an indication of the extent to which the genetics of bin variation might be explained by one or a few loci. Variation in compounds related to aromatic amino acids, branched-chain amino acids, sucrose-derived metabolites, secondary metabolites and some unidentified compounds was associated with one or more genetic loci. The combined analysis shows that there are multiple loci throughout the genome that together impact on the abundance of many compounds through a network of interactions, where individual loci may affect more than one compound and vice versa. This work therefore provides a framework for the genetic analysis of the seed metabolome, and the use of genetic marker data in the breeding and selection of seeds for specific seed quality traits and compounds that have high commercial value.
Keywords: genetic map; genetic variation; metabolite; nuclear magnetic resonance; pea; seed.
Figures




References
-
- Aubert G., Morin J., Jacquin F., Loridon K., Quillet M. C., Petit A., et al. . (2006). Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor. Appl. Genet. 112, 1024–1041. 10.1007/s00122-005-0205-y - DOI - PubMed
-
- Belton P. S., Ratcliffe R. G. (1985). NMR and compartmentation in biological tissues. Prog. Nucl. Mag. Res. Sp. 17, 241–279. 10.1016/0079-6565(85)80010-8 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

