Function and Biogenesis of Lipopolysaccharides
- PMID: 30066669
- PMCID: PMC6091223
- DOI: 10.1128/ecosalplus.ESP-0001-2018
Function and Biogenesis of Lipopolysaccharides
Abstract
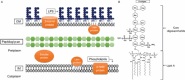
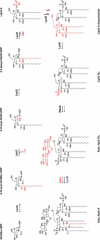
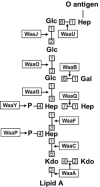
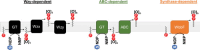
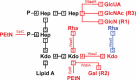
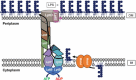
The cell envelope is the first line of defense between a bacterium and the world-at-large. Often, the initial steps that determine the outcome of chemical warfare, bacteriophage infections, and battles with other bacteria or the immune system greatly depend on the structure and composition of the bacterial cell surface. One of the most studied bacterial surface molecules is the glycolipid known as lipopolysaccharide (LPS), which is produced by most Gram-negative bacteria. Much of the initial attention LPS received in the early 1900s was owed to its ability to stimulate the immune system, for which the glycolipid was commonly known as endotoxin. It was later discovered that LPS also creates a permeability barrier at the cell surface and is a main contributor to the innate resistance that Gram-negative bacteria display against many antimicrobials. Not surprisingly, these important properties of LPS have driven a vast and still prolific body of literature for more than a hundred years. LPS research has also led to pioneering studies in bacterial envelope biogenesis and physiology, mostly using Escherichia coli and Salmonella as model systems. In this review, we will focus on the fundamental knowledge we have gained from studies of the complex structure of the LPS molecule and the biochemical pathways for its synthesis, as well as the transport of LPS across the bacterial envelope and its assembly at the cell surface.
Figures

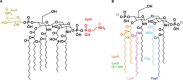
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

