A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation
- PMID: 30067292
- PMCID: PMC6585760
- DOI: 10.1111/nph.15362
A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation
Abstract
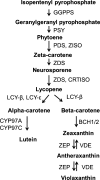
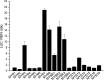
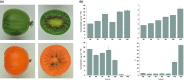
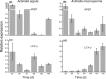
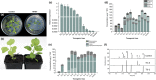
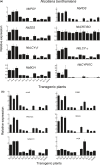
MYB transcription factors (TFs) regulate diverse plant developmental processes and understanding their roles in controlling pigment accumulation in fruit is important for developing new cultivars. In this study, we characterised kiwifruit TFMYB7, which was found to activate the promoter of the kiwifruit lycopene beta-cyclase (AdLCY-β) gene that plays a key role in the carotenoid biosynthetic pathway. To determine the role of MYB7, we analysed gene expression and metabolite profiles in Actinidia fruit which show different pigment profiles. The impact of MYB7 on metabolic biosynthetic pathways was then evaluated by overexpression in Nicotiana benthamiana followed by metabolite and gene expression analysis of the transformants. MYB7 was expressed in fruit that accumulated carotenoid and Chl pigments with high transcript levels associated with both pigments. Constitutive over-expression of MYB7, through transient or stable transformation of N. benthamiana, altered Chl and carotenoid pigment levels. MYB7 overexpression was associated with transcriptional activation of certain key genes involved in carotenoid biosynthesis, Chl biosynthesis, and other processes such as chloroplast and thylakoid membrane organization. Our results suggest that MYB7 plays a role in modulating carotenoid and Chl pigment accumulation in tissues through transcriptional activation of metabolic pathway genes.
Keywords: carotenoid; chlorophyll; kiwifruit; overexpression; transcription factor; transcriptomics.
© 2018 The Authors. New Phytologist © 2018 New Phytologist Trust.
Figures
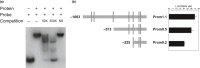

References
-
- Bou‐Torrent J, Toledo‐Ortiz G, Ortiz‐Alcaide M, Cifuentes‐Esquivel N, Halliday KJ, Martinez‐Garcia JF, Rodriguez‐Concepcion M. 2015. Regulation of carotenoid biosynthesis by shade relies on specific subsets of antagonistic transcription factors and cofactors. Plant Physiology 169: 1584–1594. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

