Autophagy Is a Tolerance-Avoidance Mechanism that Modulates TCR-Mediated Signaling and Cell Metabolism to Prevent Induction of T Cell Anergy
- PMID: 30067971
- PMCID: PMC6109966
- DOI: 10.1016/j.celrep.2018.06.065
Autophagy Is a Tolerance-Avoidance Mechanism that Modulates TCR-Mediated Signaling and Cell Metabolism to Prevent Induction of T Cell Anergy
Abstract
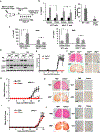
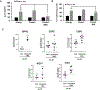
In response to activation, CD4+ T cells upregulate autophagy. However, the functional consequences of that upregulation have not been fully elucidated. In this study, we identify autophagy as a tolerance-avoidance mechanism. Our data show that inhibition of autophagy during CD4+ T cell activation induces a long-lasting state of hypo-responsiveness that is accompanied by the expression of an anergic gene signature. Cells unable to induce autophagy after T cell receptor (TCR) engagement show inefficient mitochondrial respiration and decreased turnover of the protein tyrosine phosphatase PTPN1, which translates into defective TCR-mediated signaling. In vivo, inhibition of autophagy during antigen priming induces T cell anergy and decreases the severity of disease in an experimental autoimmune encephalomyelitis mouse model. Interestingly, CD4+ T cells isolated from the synovial fluid of juvenile idiopathic arthritis patients, while resistant to suboptimal stimulation-induced anergy, can be tolerized with autophagy inhibitors. We propose that autophagy constitutes a tolerance-avoidance mechanism, which determines CD4+ T cell fate.
Keywords: T cell; anergy; autoimmunity; autophagy.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures





References
-
- Boussiotis VA, Freeman GJ, Berezovskaya A, Barber DL, and Nadler LM (1997). Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science 278, 124–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

