A map of gene expression in neutrophil-like cell lines
- PMID: 30068296
- PMCID: PMC6090850
- DOI: 10.1186/s12864-018-4957-6
A map of gene expression in neutrophil-like cell lines
Abstract
Background: Human neutrophils are central players in innate immunity, a major component of inflammatory responses, and a leading model for cell motility and chemotaxis. However, primary neutrophils are short-lived, limiting their experimental usefulness in the laboratory. Thus, human myeloid cell lines have been characterized for their ability to undergo neutrophil-like differentiation in vitro. The HL-60 cell line and its PLB-985 sub-line are commonly used to model human neutrophil behavior, but how closely gene expression in differentiated cells resembles that of primary neutrophils has remained unclear.
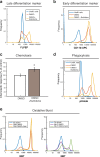
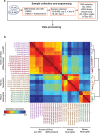
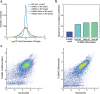
Results: In this study, we compared the effectiveness of differentiation protocols and used RNA sequencing (RNA-seq) to compare the transcriptomes of HL-60 and PLB-985 cells with published data for human and mouse primary neutrophils. Among commonly used differentiation protocols for neutrophil-like cell lines, addition of dimethyl sulfoxide (DMSO) gave the best combination of cell viability and expression of markers for differentiation. However, combining DMSO with the serum-free-supplement Nutridoma resulted in increased chemotactic response, phagocytic activity, oxidative burst and cell surface expression of the neutrophil markers FPR1 and CD11b without a cost in viability. RNA-seq analysis of HL-60 and PLB-985 cells before and after differentiation showed that differentiation broadly increases the similarity in gene expression between the cell lines and primary neutrophils. Furthermore, the gene expression pattern of the differentiated cell lines correlated slightly better with that of human neutrophils than the mouse neutrophil pattern did. Finally, we created a publicly available gene expression database that is searchable by gene name and protein domain content, where users can compare gene expression in HL-60, PLB-985 and primary human and mouse neutrophils.
Conclusions: Our study verifies that a DMSO-based differentiation protocol for HL-60 and PLB-985 cell lines gives superior differentiation and cell viability relative to other common protocols, and indicates that addition of Nutridoma may be preferable for studies of chemotaxis, phagocytosis, or oxidative burst. Our neutrophil gene expression database will be a valuable tool to identify similarities and differences in gene expression between the cell lines and primary neutrophils, to compare expression levels for genes of interest, and to improve the design of tools for genetic perturbations.
Keywords: Chemotaxis; Database; Differentiation protocol; Neutrophil; Neutrophil-like cell line; RNA-seq.
Conflict of interest statement
Ethical approval for the study of neutrophils from healthy adult volunteers was granted by the Institutional Review Board (IRB) from the University of California, Davis (IORG0000251). All participants gave written, informed consent.
Not applicable
The authors declare that they have no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

