Implications of dimeric activation of PDE6 for rod phototransduction
- PMID: 30068567
- PMCID: PMC6119862
- DOI: 10.1098/rsob.180076
Implications of dimeric activation of PDE6 for rod phototransduction
Abstract
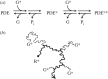
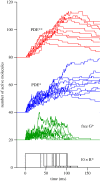
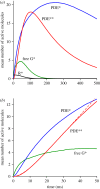
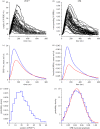
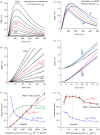
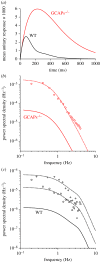
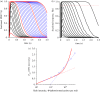
We examine the implications of a recent report providing evidence that two transducins must bind to the rod phosphodiesterase to elicit significant hydrolytic activity. To predict the rod photoreceptor's electrical response, we use numerical simulation of the two-dimensional diffusional contact of interacting molecules at the surface of the disc membrane, and then we use the simulated PDE activity as the driving function for the downstream reaction cascade. The results account for a number of aspects of rod phototransduction that have previously been puzzling. For example, they explain the existence of a greater initial delay in rods than in cones. Furthermore, our analysis suggests that the 'continuous' noise recorded in rods in darkness is likely to arise from spontaneous activation of individual molecules of PDE at a rate of a few tens per second per rod, probably as a consequence of spontaneous activation of transducins at a rate of thousands per second per rod. Hence, the dimeric activation of PDE in rods provides immunity against spontaneous transducin activation, thereby reducing the continuous noise. Our analysis also provides a coherent quantitative explanation of the amplification underlying the single photon response. Overall, numerical analysis of the dimeric activation of PDE places rod phototransduction in a new light.
Keywords: dimeric activation; phosphodiesterase PDE6; phototransduction; response kinetics; rod photoreceptors; transducin.
© 2018 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures
References
-
- Qureshi BM, et al. 2015. Asymmetric properties of rod cGMP Phosphodiesterase 6 (PDE6): structural and functional analysis. BMC Pharmacol. Toxicol. 16, 18225 (10.1186/2050-6511-16-S1-A76) - DOI
-
- Lisman J, Erickson MA, Richard EA, Cote RH, Bacigalupo J, Johnson E, Kirkwood A. 1992. Mechanisms of amplification, deactivation, and noise reduction in invertebrate photoreceptors. In Sensory transduction: Society of General Physiologists 45th annual symposium, pp. 175–199. Woods Hole, MA: Society of General Physiologists. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

