Functional Study of the C-Terminal Part of the Hepatitis C Virus E1 Ectodomain
- PMID: 30068644
- PMCID: PMC6158422
- DOI: 10.1128/JVI.00939-18
Functional Study of the C-Terminal Part of the Hepatitis C Virus E1 Ectodomain
Abstract
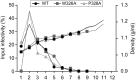
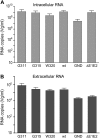
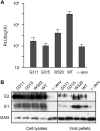
In the hepatitis C virus (HCV) envelope glycoproteins E1 and E2, which form a heterodimer, E2 is the receptor binding protein and the major target of neutralizing antibodies, whereas the function of E1 remains less characterized. To investigate E1 functions, we generated a series of mutants in the conserved residues of the C-terminal region of the E1 ectodomain in the context of an infectious clone. We focused our analyses on two regions of interest. The first region is located in the middle of the E1 glycoprotein (between amino acid [aa] 270 and aa 291), which contains a conserved hydrophobic sequence and was proposed to constitute a putative fusion peptide. The second series of mutants was generated in the region from aa 314 to aa 342 (the aa314-342 region), which has been shown to contain two α helices (α2 and α3) by nuclear magnetic resonance studies. Of the 22 generated mutants, 20 were either attenuated or noninfectious. Several mutations modulated the virus's dependence on claudin-1 and the scavenger receptor BI coreceptors for entry. Most of the mutations in the putative fusion peptide region affected virus assembly. Conversely, mutations in the α-helix aa 315 to 324 (315-324) residues M318, W320, D321, and M322 resulted in a complete loss of infectivity without any impact on E1E2 folding and on viral assembly. Further characterization of the W320A mutant in the HCVpp model indicated that the loss of infectivity was due to a defect in viral entry. Together, these results support a role for E1 in modulating HCV interaction with its coreceptors and in HCV assembly. They also highlight the involvement of α-helix 315-324 in a late step of HCV entry.IMPORTANCE HCV is a major public health problem worldwide. The virion harbors two envelope proteins, E1 and E2, which are involved at different steps of the viral life cycle. Whereas E2 has been extensively characterized, the function of E1 remains poorly defined. We characterized here the function of the putative fusion peptide and the region containing α helices of the E1 ectodomain, which had been previously suggested to be important for virus entry. We could confirm the importance of these regions for the virus infectivity. Interestingly, we found several residues modulating the virus's dependence on several HCV receptors, thus highlighting the role of E1 in the interaction of the virus with cellular receptors. Whereas mutations in the putative fusion peptide affected HCV infectivity and morphogenesis, several mutations in the α2-helix region led to a loss of infectivity with no effect on assembly, indicating a role of this region in virus entry.
Keywords: envelope proteins; glycoprotein; hepatitis C virus; viral assembly; viral entry.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
Identification of Novel Functions for Hepatitis C Virus Envelope Glycoprotein E1 in Virus Entry and Assembly.J Virol. 2017 Mar 29;91(8):e00048-17. doi: 10.1128/JVI.00048-17. Print 2017 Apr 15. J Virol. 2017. PMID: 28179528 Free PMC article.
-
Identification of conserved residues in hepatitis C virus envelope glycoprotein E2 that modulate virus dependence on CD81 and SRB1 entry factors.J Virol. 2014 Sep;88(18):10584-97. doi: 10.1128/JVI.01402-14. Epub 2014 Jul 2. J Virol. 2014. PMID: 24990994 Free PMC article.
-
Dissecting the role of putative CD81 binding regions of E2 in mediating HCV entry: putative CD81 binding region 1 is not involved in CD81 binding.Virol J. 2008 Mar 20;5:46. doi: 10.1186/1743-422X-5-46. Virol J. 2008. PMID: 18355410 Free PMC article.
-
Glycan Shielding and Modulation of Hepatitis C Virus Neutralizing Antibodies.Front Immunol. 2018 Apr 27;9:910. doi: 10.3389/fimmu.2018.00910. eCollection 2018. Front Immunol. 2018. PMID: 29755477 Free PMC article. Review.
-
Assembly of a functional HCV glycoprotein heterodimer.Curr Issues Mol Biol. 2007 Jul;9(2):71-86. Curr Issues Mol Biol. 2007. PMID: 17489436 Review.
Cited by
-
VH1-69 antiviral broadly neutralizing antibodies: genetics, structures, and relevance to rational vaccine design.Curr Opin Virol. 2019 Feb;34:149-159. doi: 10.1016/j.coviro.2019.02.004. Epub 2019 Mar 16. Curr Opin Virol. 2019. PMID: 30884330 Free PMC article. Review.
-
HCV E1 influences the fitness landscape of E2 and may enhance escape from E2-specific antibodies.Virus Evol. 2023 Nov 18;9(2):vead068. doi: 10.1093/ve/vead068. eCollection 2023. Virus Evol. 2023. PMID: 38107333 Free PMC article.
-
Prospects for developing an Hepatitis C virus E1E2-based nanoparticle vaccine.Rev Med Virol. 2023 Sep;33(5):e2474. doi: 10.1002/rmv.2474. Epub 2023 Aug 11. Rev Med Virol. 2023. PMID: 37565536 Free PMC article. Review.
-
Glycylglycine promotes the solubility and antigenic utility of recombinant HCV structural proteins in a point-of-care immunoassay for detection of active viremia.Microb Cell Fact. 2024 Jan 18;23(1):25. doi: 10.1186/s12934-024-02297-1. Microb Cell Fact. 2024. PMID: 38238770 Free PMC article.
-
Sites of vulnerability in HCV E1E2 identified by comprehensive functional screening.Cell Rep. 2022 May 24;39(8):110859. doi: 10.1016/j.celrep.2022.110859. Cell Rep. 2022. PMID: 35613596 Free PMC article.
References
-
- Simmonds P. 2013. The origin of hepatitis C virus, p 1–15. In Bartenschlager R. (ed), Hepatitis C virus: from molecular virology to antiviral therapy. Springer, Berlin, Germany.
-
- Moradpour D, Penin F. 2013. Hepatitis C virus proteins: from structure to function, p 113–142. In Bartenschlager R. (ed), Hepatitis C virus: from molecular virology to antiviral therapy. Springer, Berlin, Germany. - PubMed
-
- Cocquerel L, Wychowski C, Minner F, Penin F, Dubuisson J. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J Virol 74:3623–3633. doi:10.1128/JVI.74.8.3623-3633.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

