Mobile Genetic Elements Associated with Antimicrobial Resistance
- PMID: 30068738
- PMCID: PMC6148190
- DOI: 10.1128/CMR.00088-17
Mobile Genetic Elements Associated with Antimicrobial Resistance
Abstract
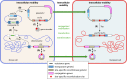
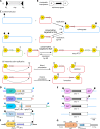
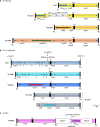
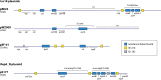
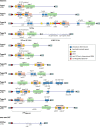
Strains of bacteria resistant to antibiotics, particularly those that are multiresistant, are an increasing major health care problem around the world. It is now abundantly clear that both Gram-negative and Gram-positive bacteria are able to meet the evolutionary challenge of combating antimicrobial chemotherapy, often by acquiring preexisting resistance determinants from the bacterial gene pool. This is achieved through the concerted activities of mobile genetic elements able to move within or between DNA molecules, which include insertion sequences, transposons, and gene cassettes/integrons, and those that are able to transfer between bacterial cells, such as plasmids and integrative conjugative elements. Together these elements play a central role in facilitating horizontal genetic exchange and therefore promote the acquisition and spread of resistance genes. This review aims to outline the characteristics of the major types of mobile genetic elements involved in acquisition and spread of antibiotic resistance in both Gram-negative and Gram-positive bacteria, focusing on the so-called ESKAPEE group of organisms (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli), which have become the most problematic hospital pathogens.
Keywords: antibiotic resistance; gene cassette; insertion sequence; integrative conjugative element; integron; plasmid; resistance island; transposon.
Copyright © 2018 American Society for Microbiology.
Figures







References
-
- Chandler M, Mahillon J. 2002. Insertion sequences revisited, p 305–366. In Craig NL, Craigie R, Gellert M, Lambowitz AM (ed), Mobile DNA II. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

