High protein copy number is required to suppress stochasticity in the cyanobacterial circadian clock
- PMID: 30068980
- PMCID: PMC6070526
- DOI: 10.1038/s41467-018-05109-4
High protein copy number is required to suppress stochasticity in the cyanobacterial circadian clock
Abstract
Circadian clocks generate reliable ~24-h rhythms despite being based on stochastic biochemical reactions. The circadian clock in Synechococcus elongatus uses a post-translational oscillator that cycles deterministically in a test tube. Because the volume of a single bacterial cell is much smaller than a macroscopic reaction, we asked how clocks in single cells function reliably. Here, we show that S. elongatus cells must express many thousands of copies of Kai proteins to effectively suppress timing errors. Stochastic modeling shows that this requirement stems from noise amplification in the post-translational feedback loop that sustains oscillations. The much smaller cyanobacterium Prochlorococcus expresses only hundreds of Kai protein copies and has a simpler, hourglass-like Kai system. We show that this timer strategy can outperform a free-running clock if internal noise is significant. This conclusion has implications for clock evolution and synthetic oscillator design, and it suggests hourglass-like behavior may be widespread in microbes.
Conflict of interest statement
The authors declare no competing interests.
Figures

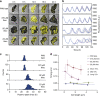
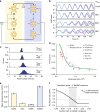
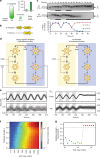
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

