Inositol phosphates are assembly co-factors for HIV-1
- PMID: 30069050
- PMCID: PMC6242333
- DOI: 10.1038/s41586-018-0396-4
Inositol phosphates are assembly co-factors for HIV-1
Erratum in
-
Author Correction: Inositol phosphates are assembly co-factors for HIV-1.Nature. 2018 Nov;563(7731):E22. doi: 10.1038/s41586-018-0505-4. Nature. 2018. PMID: 30158708
Abstract
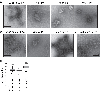
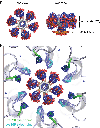
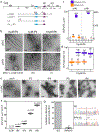
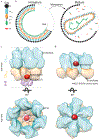
A short, 14-amino-acid segment called SP1, located in the Gag structural protein1, has a critical role during the formation of the HIV-1 virus particle. During virus assembly, the SP1 peptide and seven preceding residues fold into a six-helix bundle, which holds together the Gag hexamer and facilitates the formation of a curved immature hexagonal lattice underneath the viral membrane2,3. Upon completion of assembly and budding, proteolytic cleavage of Gag leads to virus maturation, in which the immature lattice is broken down; the liberated CA domain of Gag then re-assembles into the mature conical capsid that encloses the viral genome and associated enzymes. Folding and proteolysis of the six-helix bundle are crucial rate-limiting steps of both Gag assembly and disassembly, and the six-helix bundle is an established target of HIV-1 inhibitors4,5. Here, using a combination of structural and functional analyses, we show that inositol hexakisphosphate (InsP6, also known as IP6) facilitates the formation of the six-helix bundle and assembly of the immature HIV-1 Gag lattice. IP6 makes ionic contacts with two rings of lysine residues at the centre of the Gag hexamer. Proteolytic cleavage then unmasks an alternative binding site, where IP6 interaction promotes the assembly of the mature capsid lattice. These studies identify IP6 as a naturally occurring small molecule that promotes both assembly and maturation of HIV-1.
Figures








References
-
- Schur FK et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353, 506–508 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

