Alternative Splicing Detection Tool-a novel PERL algorithm for sensitive detection of splicing events, based on next-generation sequencing data analysis
- PMID: 30069446
- PMCID: PMC6046290
- DOI: 10.21037/atm.2018.06.32
Alternative Splicing Detection Tool-a novel PERL algorithm for sensitive detection of splicing events, based on next-generation sequencing data analysis
Abstract
Next-generation sequencing (NGS) can provide researchers with high impact information regarding alternative splice variants or transcript identifications. However, the enormous amount of data acquired from NGS platforms make the analysis of alternative splicing events hard to accomplish. For this reason, we designed the "Alternative Splicing Detection Tool" (ASDT), an algorithm that is capable of identifying alternative splicing events, including novel ones from high-throughput NGS data. ASDT is available as a PERL script at http://aias.biol.uoa.gr/~mtheo and can be executed on any system with PERL installed. In addition to the detection of annotated and novel alternative splicing events from high-throughput NGS data, ASDT can also analyze the intronic regions of genes, thus enabling the detection of novel cryptic exons residing in annotated introns, extensions of previously annotated exons, or even intron retentions. Consequently, ASDT demonstrates many innovative and unique features that can efficiently contribute to alternative splicing analysis of NGS data.
Keywords: Bioinformatics; alternative splicing; isoform; next-generation sequencing (NGS); splice variant.
Conflict of interest statement
Conflicts of Interest: The authors have no conflicts of interest to declare.
Figures
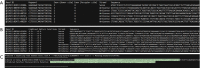
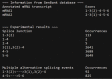
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
