Genetic Control of Myelin Plasticity after Chronic Psychosocial Stress
- PMID: 30073192
- PMCID: PMC6071195
- DOI: 10.1523/ENEURO.0166-18.2018
Genetic Control of Myelin Plasticity after Chronic Psychosocial Stress
Abstract
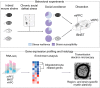
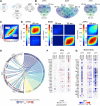
Anxiety disorders often manifest in genetically susceptible individuals after psychosocial stress, but the mechanisms underlying these gene-environment interactions are largely unknown. We used the chronic social defeat stress (CSDS) mouse model to study resilience and susceptibility to chronic psychosocial stress. We identified a strong genetic background effect in CSDS-induced social avoidance (SA) using four inbred mouse strains: 69% of C57BL/6NCrl (B6), 23% of BALB/cAnNCrl, 19% of 129S2/SvPasCrl, and 5% of DBA/2NCrl (D2) mice were stress resilient. Furthermore, different inbred mouse strains responded differently to stress, suggesting they use distinct coping strategies. To identify biological pathways affected by CSDS, we used RNA-sequencing (RNA-seq) of three brain regions of two strains, B6 and D2: medial prefrontal cortex (mPFC), ventral hippocampus (vHPC), and bed nucleus of the stria terminalis (BNST). We discovered overrepresentation of oligodendrocyte (OLG)-related genes in the differentially expressed gene population. Because OLGs myelinate axons, we measured myelin thickness and found significant region and strain-specific differences. For example, in resilient D2 mice, mPFC axons had thinner myelin than controls, whereas susceptible B6 mice had thinner myelin than controls in the vHPC. Neither myelin-related gene expression in several other regions nor corpus callosum thickness differed between stressed and control animals. Our unbiased gene expression experiment suggests that myelin plasticity is a substantial response to chronic psychosocial stress, varies across brain regions, and is genetically controlled. Identification of genetic regulators of the myelin response will provide mechanistic insight into the molecular basis of stress-related diseases, such as anxiety disorders, a critical step in developing targeted therapy.
Keywords: RNA-sequencing; anxiety; chronic social defeat stress; inbred mouse strain; myelin; transmission electron microscopy.
Figures




References
-
- Ahima RS, Harlan RE (1990) Charting of type II glucocorticoid receptor-like immunoreactivity in the rat central nervous system. Neuroscience 39:579–604. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
