SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes
- PMID: 30075142
- PMCID: PMC6095470
- DOI: 10.1016/j.molcel.2018.07.027
SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes
Abstract
Since the discovery of SUMO twenty years ago, SUMO conjugation has become a widely recognized post-translational modification that targets a myriad of proteins in many processes. Great progress has been made in understanding the SUMO pathway enzymes, substrate sumoylation, and the interplay between sumoylation and other regulatory mechanisms in a variety of contexts. As these research directions continue to generate insights into SUMO-based regulation, several mechanisms by which sumoylation and desumoylation can orchestrate large biological effects are emerging. These include the ability to target multiple proteins within the same cellular structure or process, respond dynamically to external and internal stimuli, and modulate signaling pathways involving other post-translational modifications. Focusing on nuclear function and intracellular signaling, this review highlights a broad spectrum of historical data and recent advances with the aim of providing an overview of mechanisms underlying SUMO-mediated global effects to stimulate further inquiry into intriguing roles of SUMO.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures

 . Sumoylation also regulates transcription and DNA lesion repair as indicated. Arrow: positive effects; lines: negative effects. Arrows pointing to the nuclear pore complex (NPC) and nuclear envelope indicate SUMO-mediated DNA movement toward these locations.
. Sumoylation also regulates transcription and DNA lesion repair as indicated. Arrow: positive effects; lines: negative effects. Arrows pointing to the nuclear pore complex (NPC) and nuclear envelope indicate SUMO-mediated DNA movement toward these locations.
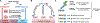
References
-
- Bachant J, Alcasabas A, Blat Y, Kleckner N, and Elledge SJ (2002). The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell 9, 1169–1182. - PubMed
-
- Baldwin ML, Julius JA, Tang XY, Wang YC, and Bachant J (2009). The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle 8, 3406–3419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

