Transcriptional activation of the miR-17-92 cluster is involved in the growth-promoting effects of MYB in human Ph-positive leukemia cells
- PMID: 30076175
- PMCID: PMC6312025
- DOI: 10.3324/haematol.2018.191213
Transcriptional activation of the miR-17-92 cluster is involved in the growth-promoting effects of MYB in human Ph-positive leukemia cells
Abstract
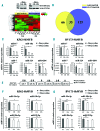
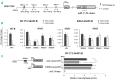
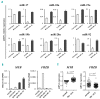
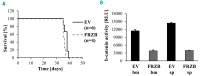
MicroRNAs, non-coding regulators of gene expression, are likely to function as important downstream effectors of many transcription factors including MYB. Optimal levels of MYB are required for transformation/maintenance of BCR-ABL-expressing cells. We investigated whether MYB silencing modulates microRNA expression in Philadelphia-positive (Ph+) leukemia cells and if MYB-regulated microRNAs are important for the "MYB addiction" of these cells. Thirty-five microRNAs were modulated by MYB silencing in lymphoid and erythromyeloid chronic myeloid leukemia-blast crisis BV173 and K562 cells; 15 of these were concordantly modulated in both lines. We focused on the miR-17-92 cluster because of its oncogenic role in tumors and found that: i) it is a direct MYB target; ii) it partially rescued the impaired proliferation and enhanced apoptosis of MYB-silenced BV173 cells. Moreover, we identified FRZB, a Wnt/β-catenin pathway inhibitor, as a novel target of the miR-17-92 cluster. High expression of MYB in blast cells from 2 Ph+leukemia patients correlated positively with the miR-17-92 cluster and inversely with FRZB. This expression pattern was also observed in a microarray dataset of 122 Ph+acute lymphoblastic leukemias. In vivo experiments in NOD scid gamma mice injected with BV173 cells confirmed that FRZB functions as a Wnt/β-catenin inhibitor even as they failed to demonstrate that this pathway is important for BV173-dependent leukemogenesis. These studies illustrate the global effects of MYB expression on the microRNAs profile of Ph+cells and supports the concept that the "MYB addiction" of these cells is, in part, caused by modulation of microRNA-regulated pathways affecting cell proliferation and survival.
Copyright© 2019 Ferrata Storti Foundation.
Figures


References
-
- Perrotti D, Cesi V, Trotta R, et al. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat Genet. 2002;30(1):48–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

