Fitting mathematical models of biochemical pathways to steady state perturbation response data without simulating perturbation experiments
- PMID: 30076370
- PMCID: PMC6076289
- DOI: 10.1038/s41598-018-30118-0
Fitting mathematical models of biochemical pathways to steady state perturbation response data without simulating perturbation experiments
Abstract
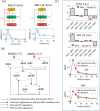
Fitting Ordinary Differential Equation (ODE) models of signal transduction networks (STNs) to experimental data is a challenging problem. Computational parameter fitting algorithms simulate a model many times with different sets of parameter values until the simulated STN behaviour match closely with experimental data. This process can be slow when the model is fitted to measurements of STN responses to numerous perturbations, since this requires simulating the model as many times as the number of perturbations for each set of parameter values. Here, I propose an approach that avoids simulating perturbation experiments when fitting ODE models to steady state perturbation response (SSPR) data. Instead of fitting the model directly to SSPR data, it finds model parameters which provides a close match between the scaled Jacobian matrices (SJM) of the model, which are numerically calculated using the model's rate equations and estimated from SSPR data using modular response analysis (MRA). The numerical estimation of SJM of an ODE model does not require simulating perturbation experiments, saving significant computation time. The effectiveness of this approach is demonstrated by fitting ODE models of the Mitogen Activated Protein Kinase (MAPK) pathway using simulated and real SSPR data.
Conflict of interest statement
The author declares no competing interests.
Figures




References
-
- Girolami M, Calderhead B, Girolami M, Calderhead B. Riemann manifold Langevin and Hamiltonian Monte Carlo methods. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2011;73:123–214. doi: 10.1111/j.1467-9868.2010.00765.x. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

