Pyrophosphorylation via selective phosphoprotein derivatization
- PMID: 30079207
- PMCID: PMC6050540
- DOI: 10.1039/c8sc01233d
Pyrophosphorylation via selective phosphoprotein derivatization
Abstract
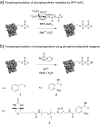
An important step in elucidating the function of protein post-translational modifications (PTMs) is gaining access to site-specifically modified, homogeneous samples for biochemical characterization. Protein pyrophosphorylation is a poorly characterized PTM, and here a chemical approach to obtain pyrophosphoproteins is reported. Photo-labile phosphorimidazolide reagents were developed for selective pyrophosphorylation, affinity-capture, and release of pyrophosphoproteins. Kinetic analysis of the reaction revealed rate constants between 9.2 × 10-3 to 0.58 M-1 s-1, as well as a striking proclivity of the phosphorimidazolides to preferentially react with phosphate monoesters over other nucleophilic side chains. Besides enabling the characterization of pyrophosphorylation on protein function, this work highlights the utility of phosphoryl groups as handles for selective protein modification for a variety of applications, such as phosphoprotein bioconjugation and enrichment.
Figures






References
-
- Walsh C. T., Posttranslational Modifications of Proteins: Expanding Nature's Inventory, Roberts and Company Publishers, Greenwood Village, Colorado, 2006.
-
- Attwood P. V., Piggott M. J., Zu X. L., Besant P. G. Amino Acids. 2007;32:145–156. - PubMed
-
- Besant P. G., Attwood P. V., Piggott M. J. Curr. Protein Pept. Sci. 2009;10:536–550. - PubMed
-
- Attwood P. V., Besant P. G., Piggott M. J. Amino Acids. 2011;40:1035–1051. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

