Distinct transcriptional responses of mouse sensory neurons in models of human chronic pain conditions
- PMID: 30079380
- PMCID: PMC6053702
- DOI: 10.12688/wellcomeopenres.14641.1
Distinct transcriptional responses of mouse sensory neurons in models of human chronic pain conditions
Abstract
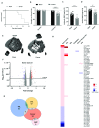
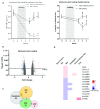
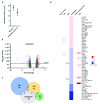
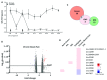
Background: Sensory neurons play an essential role in almost all pain conditions, and have recently been classified into distinct subsets on the basis of their transcriptomes. Here we have analysed alterations in dorsal root ganglia (DRG) gene expression using microarrays in mouse models related to human chronic pain. Methods: Six different pain models were studied in male C57BL/6J mice: (1) bone cancer pain using cancer cell injection in the intramedullary space of the femur; (2) neuropathic pain using partial sciatic nerve ligation; (3) osteoarthritis pain using mechanical joint loading; (4) chemotherapy-induced pain with oxaliplatin; (5) chronic muscle pain using hyperalgesic priming; and (6) inflammatory pain using intraplantar complete Freund's adjuvant. Microarray analyses were performed using RNA isolated from dorsal root ganglia and compared to sham/vehicle treated controls. Results: Differentially expressed genes (DEGs) were identified. Known and previously unreported genes were found to be dysregulated in each pain model. The transcriptomic profiles for each model were compared and expression profiles of DEGs within subsets of DRG neuronal populations were analysed to determine whether specific neuronal subsets could be linked to each of the pain models. Conclusions: Each pain model exhibits a unique set of altered transcripts implying distinct cellular responses to different painful stimuli. No simple direct link between genetically distinct sets of neurons and particular pain models could be discerned.
Keywords: Chronic pain; dorsal root ganglia; gene expression; microarrays; mouse models; sensory neurons.
Conflict of interest statement
No competing interests were disclosed.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

