Emerging techniques in single-cell epigenomics and their applications to cancer research
- PMID: 30079405
- PMCID: PMC6070152
- DOI: 10.4172/JCG.1000103
Emerging techniques in single-cell epigenomics and their applications to cancer research
Abstract
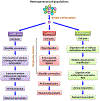
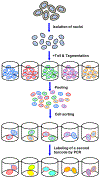
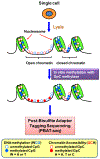
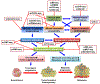
Epigenomics encompasses studies of the chemical modifications of genomic DNA and associated histones, interactions between genomic DNA sequences and proteins, the dynamics of the chromosomal conformation, the functional relationships between these epigenetic events, and the regulatory impacts of these epigenetic events on gene expression in cells. In comparison to current techniques that are only capable of characterizing average epigenomic features across bulk cell ensembles, single-cell epigenomic methodologies are emerging as powerful new techniques to study cellular plasticity and heterogeneity, as seen in stem cells and cancer. Here we summarize available techniques for studies of single-cell epigenomics, review their current applications to cancer research, and discuss future possibilities. This review also highlights that the full potential of single-cell epigenetic studies will be comprehended through integrating the multi-omics information of genomics, epigenomics and transcriptomics.
Keywords: Chemotherapeutic treatments; Epigenomics; Genomic DNA; Oncogenesis; intratumor heterogeneity; single-cell techniques.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
