Identification of Functional Variants in the FAM13A Chronic Obstructive Pulmonary Disease Genome-Wide Association Study Locus by Massively Parallel Reporter Assays
- PMID: 30079747
- PMCID: PMC6353020
- DOI: 10.1164/rccm.201802-0337OC
Identification of Functional Variants in the FAM13A Chronic Obstructive Pulmonary Disease Genome-Wide Association Study Locus by Massively Parallel Reporter Assays
Abstract
Rationale: The identification of causal variants responsible for disease associations from genome-wide association studies (GWASs) facilitates functional understanding of the biological mechanisms by which those genetic variants influence disease susceptibility.
Objective: We aim to identify causal variants in or near the FAM13A (family with sequence similarity member 13A) GWAS locus associated with chronic obstructive pulmonary disease (COPD).
Methods: We used an integrated approach featuring conditional genetic analysis, massively parallel reporter assays (MPRAs), traditional reporter assays, chromatin conformation capture assays, and clustered regularly interspaced short palindromic repeats (CRISPR)-based gene editing to characterize COPD-associated regulatory variants in the FAM13A region in human bronchial epithelial cell lines.
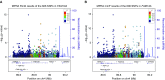
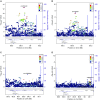
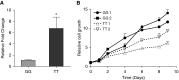
Measurements and main results: Conditional genetic association suggests the presence of two independent COPD association signals in FAM13A. MPRAs identified 45 regulatory variants within FAM13A, among which six variants were prioritized for further investigation. Three COPD-associated variants demonstrated significant allele-specific activity in reporter assays. One of three variants, rs2013701, was tested in the endogenous genomic context by CRISPR-based genome editing that confirmed its allele-specific effects on FAM13A expression and on cell proliferation, providing functional characterization for this COPD-associated variant.
Conclusions: The human GWAS association near FAM13A may contain independent association signals. MPRAs identified multiple functional variants in this region, including rs2013701, a putative COPD-causing variant with allele-specific regulatory activity.
Keywords: CRISPR editing; chromosome conformation capture; functional genomics; human bronchial epithelial cells; reporter assay.
Figures



Comment in
-
Deciphering the Genetics of Chronic Obstructive Pulmonary Disease.Am J Respir Crit Care Med. 2019 Jan 1;199(1):4-5. doi: 10.1164/rccm.201808-1465ED. Am J Respir Crit Care Med. 2019. PMID: 30130126 No abstract available.
References
-
- Hobbs BD, de Jong K, Lamontagne M, Bossé Y, Shrine N, Artigas MS, et al. COPDGene Investigators; ECLIPSE Investigators; LifeLines Investigators; SPIROMICS Research Group; International COPD Genetics Network Investigators; UK BiLEVE Investigators; International COPD Genetics Consortium. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49:426–432. - PMC - PubMed
-
- Skronska-Wasek W, Mutze K, Baarsma HA, Bracke KR, Alsafadi HN, Lehmann M, et al. Reduced frizzled receptor 4 expression prevents WNT/β-catenin-driven alveolar lung repair in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017;196:172–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL113264/HL/NHLBI NIH HHS/United States
- R01 HL135142/HL/NHLBI NIH HHS/United States
- R01 HL126596/HL/NHLBI NIH HHS/United States
- K01 HL129039/HL/NHLBI NIH HHS/United States
- R01 HL124233/HL/NHLBI NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- R33 HL120794/HL/NHLBI NIH HHS/United States
- P01 HL105339/HL/NHLBI NIH HHS/United States
- R01 HG006785/HG/NHGRI NIH HHS/United States
- R01 HL137927/HL/NHLBI NIH HHS/United States
- R01 HL127200/HL/NHLBI NIH HHS/United States
- P01 HL114501/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

