Modeling of the GC content of the substituted bases in bacterial core genomes
- PMID: 30081825
- PMCID: PMC6080486
- DOI: 10.1186/s12864-018-4984-3
Modeling of the GC content of the substituted bases in bacterial core genomes
Abstract
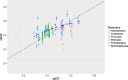
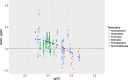
Background: The purpose of the present study was to examine the GC content of substituted bases (sbGC) in the core genomes of 35 bacterial species. Each species, or core genome, constituted genomes from at least 10 strains. We also wanted to explore whether sbGC for each strain was associated with the corresponding species' core genome GC content (cgGC). We present a simple mathematical model that estimates sbGC from cgGC. The model assumes only that the estimated sbGC is a function of cgGC proportional to fixed AT→GC (α) and GC → AT (β) mutation rates. Non-linear regression was used to estimate parameters α and β from the empirical data described above.
Results: We found that sbGC for each strain showed a non-linear association with the corresponding cgGC with a bias towards higher GC content for most core genomes (66.3% of the strains), assuming as a null-hypothesis that sbGC should be approximately equal to cgGC. The most GC rich core genomes (i.e. approximately %GC > 60), on the other hand, exhibited slightly less GC-biased sbGC than expected. The best fitted regression model indicates that GC → AT mutation rates β = (1.91 ± 0.13) p < 0.001 are approximately (1.91/0.79) = 2.42 times as high, on average, as AT→GC α = (- 0.79 ± 0.25) p < 0.001 mutation rates. Whether the observed sbGC GC-bias for all but the most GC-rich prokaryotic species is due to selection, compensating for the GC → AT mutation bias, and/or selective neutral processes is currently debated. Residual standard error was found to be σ = 0.076 indicating estimated errors of sbGC to be approximately within ±15.2% GC (95% confidence interval) for the strains of all species in the study.
Conclusion: Not only did our mathematical model give reasonable estimates of sbGC it also provides further support to previous observations that mutation rates in prokaryotes exhibit a universal GC → AT bias that appears to be remarkably consistent between taxa.
Keywords: Core genome; Core genome GC content; Mathematical modeling; Microbial genomics; SNP GC content; Statistical parameter estimation.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

