MuscleJ: a high-content analysis method to study skeletal muscle with a new Fiji tool
- PMID: 30081940
- PMCID: PMC6091189
- DOI: 10.1186/s13395-018-0171-0
MuscleJ: a high-content analysis method to study skeletal muscle with a new Fiji tool
Abstract
Background: Skeletal muscle has the capacity to adapt to environmental changes and regenerate upon injury. To study these processes, most experimental methods use quantification of parameters obtained from images of immunostained skeletal muscle. Muscle cross-sectional area, fiber typing, localization of nuclei within the muscle fiber, the number of vessels, and fiber-associated stem cells are used to assess muscle physiology. Manual quantification of these parameters is time consuming and only poorly reproducible. While current state-of-the-art software tools are unable to analyze all these parameters simultaneously, we have developed MuscleJ, a new bioinformatics tool to do so.
Methods: Running on the popular open source Fiji software platform, MuscleJ simultaneously analyzes parameters from immunofluorescent staining, imaged by different acquisition systems in a completely automated manner.
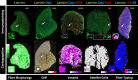
Results: After segmentation of muscle fibers, up to three other channels can be analyzed simultaneously. Dialog boxes make MuscleJ easy-to-use for biologists. In addition, we have implemented color in situ cartographies of results, allowing the user to directly visualize results on reconstituted muscle sections.
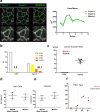
Conclusion: We report here that MuscleJ results were comparable to manual observations made by five experts. MuscleJ markedly enhances statistical analysis by allowing reliable comparison of skeletal muscle physiology-pathology results obtained from different laboratories using different acquisition systems. Providing fast robust multi-parameter analyses of skeletal muscle physiology-pathology, MuscleJ is available as a free tool for the skeletal muscle community.
Keywords: Fiber typing; Histology; Image automated quantification; In situ cartography; Satellite cells; Skeletal muscle fiber; Stem cells; Vessels.
Conflict of interest statement
Experimental procedures were performed with the approval of the Nord-Pas-de-Calais ethics committee (CEEA75- 2017122019046377), the Institut Pasteur Ethics Committee (C2EA 89 - CETEA), and in compliance with French and European ethical legislations. Protocols were also reviewed by the Institut Pasteur, the competent authority, for compliance with the French and European regulations on Animal Welfare and with Public Health Service recommendations. This project has been reviewed and approved (# 2013-0044) by the Institut Pasteur Ethics Committee (C2EA 89 - CETEA).
Not applicable.
The authors declare that they have no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Buckingham M, Mayeuf A. Skeletal muscle development. In: Hill JA, Olson EN, editors. Muscle: fundamental biology and mechanisms of disease. Boston: Academic Press; 2012. p. 749-62.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

