Structure of the EmrE multidrug transporter and its use for inhibitor peptide design
- PMID: 30082384
- PMCID: PMC6112734
- DOI: 10.1073/pnas.1802177115
Structure of the EmrE multidrug transporter and its use for inhibitor peptide design
Abstract
Small multidrug resistance (SMR) pumps represent a minimal paradigm of proton-coupled membrane transport in bacteria, yet no high-resolution structure of an SMR protein is available. Here, atomic-resolution structures of the Escherichia coli efflux-multidrug resistance E (EmrE) multidrug transporter in ligand-bound form are refined using microsecond molecular dynamics simulations biased using low-resolution data from X-ray crystallography. The structures are compatible with existing mutagenesis data as well as NMR and biochemical experiments, including pKas of the catalytic glutamate residues and the dissociation constant ([Formula: see text]) of the tetraphenylphosphonium+ cation. The refined structures show the arrangement of residue side chains in the EmrE active site occupied by two different ligands and in the absence of a ligand, illustrating how EmrE can adopt structurally diverse active site configurations. The structures also show a stable, well-packed binding interface between the helices H4 of the two monomers, which is believed to be crucial for EmrE dimerization. Guided by the atomic details of this interface, we design proteolysis-resistant stapled peptides that bind to helix H4 of an EmrE monomer. The peptides are expected to interfere with the dimerization and thereby inhibit drug transport. Optimal positions of the peptide staple were determined using free-energy simulations of peptide binding to monomeric EmrE Three of the four top-scoring peptides selected for experimental testing resulted in significant inhibition of proton-driven ethidium efflux in live cells without nonspecific toxicity. The approach described here is expected to be of general use for the design of peptide therapeutics.
Keywords: drug resistance; membrane proteins; molecular dynamics; stapled peptides; structure refinement.
Figures





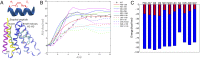
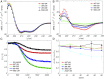
References
-
- Bay D, Rommens K, Turner R. Small multidrug resistance proteins: A multidrug transporter family that continues to grow. Biochim Biophys Acta. 2008;1778:1814–1838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

