Epigenetic control of innate and adaptive immune memory
- PMID: 30082830
- PMCID: PMC6225771
- DOI: 10.1038/s41590-018-0176-1
Epigenetic control of innate and adaptive immune memory
Abstract
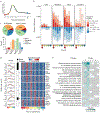
Clonal expansion and immunological memory are hallmark features of the mammalian adaptive immune response and essential for prolonged host control of pathogens. Recent work demonstrates that natural killer (NK) cells of the innate immune system also exhibit these adaptive traits during infection. Here we demonstrate that differentiating and 'memory' NK cells possess distinct chromatin accessibility states and that their epigenetic profiles reveal a 'poised' regulatory program at the memory stage. Furthermore, we elucidate how individual STAT transcription factors differentially control epigenetic and transcriptional states early during infection. Finally, concurrent chromatin profiling of the canonical CD8+ T cell response against the same infection demonstrated parallel and distinct epigenetic signatures defining NK cells and CD8+ T cells. Overall, our study reveals the dynamic nature of epigenetic modifications during the generation of innate and adaptive lymphocyte memory.
Figures



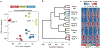

Comment in
-
Inscribing the core memories of killers.Cell Mol Immunol. 2019 Feb;16(2):104-105. doi: 10.1038/s41423-018-0178-9. Epub 2018 Nov 7. Cell Mol Immunol. 2019. PMID: 30405147 Free PMC article. No abstract available.
-
Warm up, cool down, and tearing apart in NK cell memory.Cell Mol Immunol. 2018 Dec;15(12):1095-1097. doi: 10.1038/s41423-018-0188-7. Epub 2018 Nov 28. Cell Mol Immunol. 2018. PMID: 30487549 Free PMC article. No abstract available.
References
-
- Arase H, Mocarski ES, Campbell AE, Hill AB & Lanier LL Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296, 1323–1326 (2002). - PubMed
-
- Dokun AO et al. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol 2, 951–956 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

