Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography
- PMID: 30082915
- PMCID: PMC6119093
- DOI: 10.1038/s41593-018-0195-0
Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography
Abstract
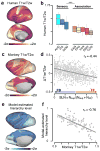
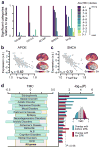
Hierarchy provides a unifying principle for the macroscale organization of anatomical and functional properties across primate cortex, yet microscale bases of specialization across human cortex are poorly understood. Anatomical hierarchy is conventionally informed by invasive tract-tracing measurements, creating a need for a principled proxy measure in humans. Moreover, cortex exhibits marked interareal variation in gene expression, yet organizing principles of cortical transcription remain unclear. We hypothesized that specialization of cortical microcircuitry involves hierarchical gradients of gene expression. We found that a noninvasive neuroimaging measure-MRI-derived T1-weighted/T2-weighted (T1w/T2w) mapping-reliably indexes anatomical hierarchy, and it captures the dominant pattern of transcriptional variation across human cortex. We found hierarchical gradients in expression profiles of genes related to microcircuit function, consistent with monkey microanatomy, and implicated in neuropsychiatric disorders. Our findings identify a hierarchical axis linking cortical transcription and anatomy, along which gradients of microscale properties may contribute to the macroscale specialization of cortical function.
Conflict of interest statement
The authors declare the following competing interests: This research was partly funded by BlackThorn Therapeutics. W.J.M. is an employee for BlackThorn Therapeutics. A.A. and J.D.M. are consultants for BlackThorn Therapeutics. W.J.M., A.A., and J.D.M. are co-inventors on a provisional patent application #62567087 related to using gene expression topography for predictive therapeutic applications.
Figures


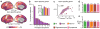


References
-
- Lennie P. Single units and visual cortical organization. Perception. 1998;27:889–935. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

