An efficient CRISPR vector toolbox for engineering large deletions in Arabidopsis thaliana
- PMID: 30083222
- PMCID: PMC6071326
- DOI: 10.1186/s13007-018-0330-7
An efficient CRISPR vector toolbox for engineering large deletions in Arabidopsis thaliana
Abstract
Background: Our knowledge of natural genetic variation is increasing at an extremely rapid pace, affording an opportunity to come to a much richer understanding of how effects of specific genes are dependent on the genetic background. To achieve a systematic understanding of such GxG interactions, it is desirable to develop genome editing tools that can be rapidly deployed across many different genetic varieties.
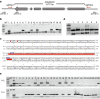
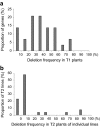
Results: We present an efficient CRISPR/Cas9 toolbox of super module (SM) vectors. These vectors are based on a previously described fluorescence protein marker expressed in seeds allowing identification of transgene-free mutants. We have used this vector series to delete genomic regions ranging from 1.7 to 13 kb in different natural accessions of the wild plant Arabidopsis thaliana. Based on results from 53 pairs of sgRNAs targeting individual nucleotide binding site leucine-rich repeat (NLR) genes, we provide a comprehensive overview of obtaining heritable deletions.
Conclusions: The SM series of CRISPR/Cas9 vectors enables the rapid generation of transgene-free, genome edited plants for a diversity of functional studies.
Keywords: Arabidopsis thaliana; CRISPR/Cas9; Genome editing; NLR genes; Natural variation.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

