Review of applications of high-throughput sequencing in personalized medicine: barriers and facilitators of future progress in research and clinical application
- PMID: 30084865
- PMCID: PMC6917217
- DOI: 10.1093/bib/bby051
Review of applications of high-throughput sequencing in personalized medicine: barriers and facilitators of future progress in research and clinical application
Abstract
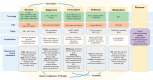
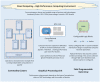
There has been an exponential growth in the performance and output of sequencing technologies (omics data) with full genome sequencing now producing gigabases of reads on a daily basis. These data may hold the promise of personalized medicine, leading to routinely available sequencing tests that can guide patient treatment decisions. In the era of high-throughput sequencing (HTS), computational considerations, data governance and clinical translation are the greatest rate-limiting steps. To ensure that the analysis, management and interpretation of such extensive omics data is exploited to its full potential, key factors, including sample sourcing, technology selection and computational expertise and resources, need to be considered, leading to an integrated set of high-performance tools and systems. This article provides an up-to-date overview of the evolution of HTS and the accompanying tools, infrastructure and data management approaches that are emerging in this space, which, if used within in a multidisciplinary context, may ultimately facilitate the development of personalized medicine.
Keywords: clinical translation; cloud computing; grid computing; high-performance computing; high-throughput sequencing; personalized medicine; translational research.
© The Author(s) 2018. Published by Oxford University Press.
Figures

Similar articles
-
FDA's Activities Supporting Regulatory Application of "Next Gen" Sequencing Technologies.PDA J Pharm Sci Technol. 2014 Nov-Dec;68(6):626-30. doi: 10.5731/pdajpst.2014.01024. PDA J Pharm Sci Technol. 2014. PMID: 25475637
-
Bioinformatics and Microarray Data Analysis on the Cloud.Methods Mol Biol. 2016;1375:25-39. doi: 10.1007/7651_2015_236. Methods Mol Biol. 2016. PMID: 25863787
-
An Overview of DNA Analytical Methods.Methods Mol Biol. 2019;1897:385-402. doi: 10.1007/978-1-4939-8935-5_31. Methods Mol Biol. 2019. PMID: 30539459 Review.
-
Explanatory chapter: next generation sequencing.Methods Enzymol. 2013;529:201-8. doi: 10.1016/B978-0-12-418687-3.00016-1. Methods Enzymol. 2013. PMID: 24011047 Free PMC article.
-
Implementation of cancer next-generation sequencing testing in a community hospital.Cold Spring Harb Mol Case Stud. 2019 Jun 3;5(3):a003707. doi: 10.1101/mcs.a003707. Print 2019 Jun. Cold Spring Harb Mol Case Stud. 2019. PMID: 31160354 Free PMC article.
Cited by
-
Comparative whole transcriptome analysis of gene expression in three canine soft tissue sarcoma types.PLoS One. 2022 Sep 13;17(9):e0273705. doi: 10.1371/journal.pone.0273705. eCollection 2022. PLoS One. 2022. PMID: 36099287 Free PMC article.
-
Cutaneous Melanoma Classification: The Importance of High-Throughput Genomic Technologies.Front Oncol. 2021 May 28;11:635488. doi: 10.3389/fonc.2021.635488. eCollection 2021. Front Oncol. 2021. PMID: 34123788 Free PMC article. Review.
-
Interpretable deep neural network for cancer survival analysis by integrating genomic and clinical data.BMC Med Genomics. 2019 Dec 23;12(Suppl 10):189. doi: 10.1186/s12920-019-0624-2. BMC Med Genomics. 2019. PMID: 31865908 Free PMC article.
-
Comprehensive genomics analysis of aging related gene signature to predict the prognosis and drug resistance of colon adenocarcinoma.Front Pharmacol. 2023 Feb 28;14:1121634. doi: 10.3389/fphar.2023.1121634. eCollection 2023. Front Pharmacol. 2023. PMID: 36925638 Free PMC article.
-
Epione application: An integrated web‑toolkit of clinical genomics and personalized medicine in systemic lupus erythematosus.Int J Mol Med. 2022 Jan;49(1):8. doi: 10.3892/ijmm.2021.5063. Epub 2021 Nov 18. Int J Mol Med. 2022. PMID: 34791504 Free PMC article.
References
-
- Illumina. NovaSeq series—the next era in sequencing starts now. Illumina, 2018. https://www.illumina.com/systems/sequencing-platforms/novaseq.html
-
- Fikes B. New machines can sequence human genome in one hour, Illumina announces. The San Diego Union-Tribune. 2017. http://www.sandiegouniontribune.com/business/biotech/sd-me-illumina-nova...
-
- Edico Genome. DRAGEN Bio-IT platform. Edico Genome, 2018. http://edicogenome.com/dragen-bioit-platform/
-
- Baker M. Next-generation sequencing: adjusting to data overload. Nat Methods 2010;7(7):495–9.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

