Hot-starting software containers for STAR aligner
- PMID: 30085034
- PMCID: PMC6131214
- DOI: 10.1093/gigascience/giy092
Hot-starting software containers for STAR aligner
Abstract
Background: Using software containers has become standard practice to reproducibly deploy and execute biomedical workflows on the cloud. However, some applications that contain time-consuming initialization steps will produce unnecessary costs for repeated executions.
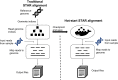
Findings: We demonstrate that hot-starting from containers that have been frozen after the application has already begun execution can speed up bioinformatics workflows by avoiding repetitive initialization steps. We use an open-source tool called Checkpoint and Restore in Userspace (CRIU) to save the state of the containers as a collection of checkpoint files on disk after it has read in the indices. The resulting checkpoint files are migrated to the host, and CRIU is used to regenerate the containers in that ready-to-run hot-start state. As a proof-of-concept example, we create a hot-start container for the spliced transcripts alignment to a reference (STAR) aligner and deploy this container to align RNA sequencing data. We compare the performance of the alignment step with and without checkpoints on cloud platforms using local and network disks.
Conclusions: We demonstrate that hot-starting Docker containers from snapshots taken after repetitive initialization steps are completed significantly speeds up the execution of the STAR aligner on all experimental platforms, including Amazon Web Services, Microsoft Azure, and local virtual machines. Our method can be potentially employed in other bioinformatics applications in which a checkpoint can be inserted after a repetitive initialization phase.
Figures

References
-
- Calabrese B, Cannataro M. Cloud computing in bioinformatics: current solutions and challenges. Peer J Preprints. 2016;4:e2261v1.
-
- National Institutes of Health (NIH) Data Commons Pilot. https://commonfund.nih.gov/commons. Accessed 6 April 2018.
-
- Docker. https://http://www.docker.com/. Accessed 6 April 2018.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

