Biased Inference of Selection Due to GC-Biased Gene Conversion and the Rate of Protein Evolution in Flycatchers When Accounting for It
- PMID: 30085180
- PMCID: PMC6188562
- DOI: 10.1093/molbev/msy149
Biased Inference of Selection Due to GC-Biased Gene Conversion and the Rate of Protein Evolution in Flycatchers When Accounting for It
Abstract
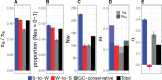
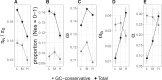
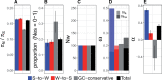
The rate of recombination impacts on rates of protein evolution for at least two reasons: it affects the efficacy of selection due to linkage and influences sequence evolution through the process of GC-biased gene conversion (gBGC). We studied how recombination, via gBGC, affects inferences of selection in gene sequences using comparative genomic and population genomic data from the collared flycatcher (Ficedula albicollis). We separately analyzed different mutation categories ("strong"-to-"weak," "weak-to-strong," and GC-conservative changes) and found that gBGC impacts on the distribution of fitness effects of new mutations, and leads to that the rate of adaptive evolution and the proportion of adaptive mutations among nonsynonymous substitutions are underestimated by 22-33%. It also biases inferences of demographic history based on the site frequency spectrum. In light of this impact, we suggest that inferences of selection (and demography) in lineages with pronounced gBGC should be based on GC-conservative changes only. Doing so, we estimate that 10% of nonsynonymous mutations are effectively neutral and that 27% of nonsynonymous substitutions have been fixed by positive selection in the flycatcher lineage. We also find that gene expression level, sex-bias in expression, and the number of protein-protein interactions, but not Hill-Robertson interference (HRI), are strong determinants of selective constraint and rate of adaptation of collared flycatcher genes. This study therefore illustrates the importance of disentangling the effects of different evolutionary forces and genetic factors in interpretation of sequence data, and from that infer the role of natural selection in DNA sequence evolution.
Figures
References
-
- Axelsson E, Ellegren H. 2009. Quantification of adaptive evolution of genes expressed in avian brain and the population size effect on the efficacy of selection. Mol Biol Evol. 265:1073–1079. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

