Mucosal Gene Expression in Pediatric and Adult Patients With Ulcerative Colitis Permits Modeling of Ideal Biopsy Collection Strategy for Transcriptomic Analysis
- PMID: 30085215
- PMCID: PMC6265054
- DOI: 10.1093/ibd/izy242
Mucosal Gene Expression in Pediatric and Adult Patients With Ulcerative Colitis Permits Modeling of Ideal Biopsy Collection Strategy for Transcriptomic Analysis
Abstract
Background: Transcriptional profiling has been performed on biopsies from ulcerative colitis patients. Limitations in prior studies include the variability introduced by inflammation, anatomic site of biopsy, extent of disease, and medications. We sought to more globally understand the variability of gene expression from patients with ulcerative colitis to advance our understanding of its pathogenesis and to guide clinical study design.
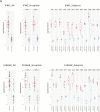
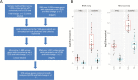
Methods: We performed transcriptional profiling on 13 subjects, including pediatric and adult patients from 2 hospital sites. For each patient, we collected 6 biopsies from macroscopically inflamed tissue and 4 biopsies from macroscopically healthy-appearing tissue. Isolated RNA was used for microarray gene expression analysis utilizing Affymetrix Human Primeview microarrays. Ingenuity pathway analysis was used to assess over-representation of gene ontology and biological pathways. RNAseq was also performed, and differential analysis was assessed to compare affected vs unaffected samples. Finally, we modeled the minimum number of biopsies required to reliably detect gene expression across different subject numbers.
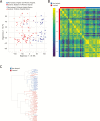
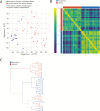
Results: Transcriptional profiles co-clustered independently of the hospital collection site, patient age, sex, and colonic location, which parallels prior gene expression findings. A small set of genes not previously described was identified. Our modeling analysis reveals the number of biopsies and patients per cohort to yield reliable results in clinical studies.
Conclusions: Key findings include concordance, including some expansion, of previously published gene expression studies and similarity among different age groups. We also established a reliable statistical model for biopsy collection for future clinical studies.
Figures

References
-
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

