Transcriptome analysis of rhesus monkey failed-to-mature oocytes: deficiencies in transcriptional regulation and cytoplasmic maturation of the oocyte mRNA population
- PMID: 30085220
- PMCID: PMC6497044
- DOI: 10.1093/molehr/gay032
Transcriptome analysis of rhesus monkey failed-to-mature oocytes: deficiencies in transcriptional regulation and cytoplasmic maturation of the oocyte mRNA population
Abstract
Study question: Which different pathways and functions are altered in rhesus monkey oocytes that fail to mature after an ovulatory stimulus?
Summary answer: Failed to mature (FTM) oocytes complete a large portion of the transition in transcriptome composition associated with normal maturation, but also manifest numerous differences that indicate incomplete transcriptional repression and cytoplasmic maturation affecting multiple processes.
What is known already: Oocyte maturation defects contribute to unexplained female infertility. Failure of some oocytes to undergo germinal vesicle breakdown or progress to second meiotic metaphase in response to an ovulatory stimulus can limit the number of high quality oocytes available for ART.
Study design, size, duration: The transcriptome of rhesus monkey oocytes that failed to mature (FTM; n = 11, 5 donors) in response to an ovulatory stimulus in vivo was compared to those of normal germinal vesicle stage (GV, n = 7, 2 donors) and metaphase II stage (MII, n = 7, 5 donors) oocytes by RNA-sequencing (RNAseq).
Participants/materials, setting, methods: Female rhesus monkeys of normal breeding age (6-12 years old) and with regular menstrual cycles were used. Animals underwent a controlled ovarian stimulation protocol for the collection of oocytes by ultrasound-guided needle aspiration of follicles.
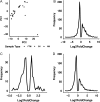
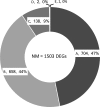
Main results and the role of chance: We obtained a high quality RNAseq dataset consisting of n = 7, n = 7, and n = 11 libraries for normal GV, normal MII and FTM oocytes, respectively. Total reads acquired were an average of 34 million for each GV sample, 41 million for each FTM sample and 59 million for each MII oocyte sample. Approximately 44% of the total reads were exonic reads that successfully aligned to the rhesus monkey genome as unique non-rRNA gene transcript sequences, providing high depth of coverage. Approximately 44% of the mRNAs that undergo changes in abundance during normal maturation display partial modulations to intermediate abundances, and 9.2% fail to diverge significantly from GV stage oocytes. Additionally, a small group of mRNAs are grossly mis-regulated in the FTM oocyte. Differential expression was seen for mRNAs associated with mitochondrial functions, fatty acid beta oxidation, lipid accumulation, meiosis, zona pellucida formation, Hippo pathway signaling, and maternal mRNA regulation. A deficiency DNA methyltransferase one mRNA expression indicates a potential defect in transcriptional silencing.
Large scale data: All RNAseq data are published in the Gene Expression Omnibus Database (GSE112536).
Limitations, reasons for caution: These results do not establish cause of maturation failure but reveal novel correlates of incompetence to mature. Transcriptome studies likely do not capture all post-transcriptional or post-translational events that inhibit maturation, but do reveal mRNA expression changes that lie downstream of such events or that are related to effects on upstream regulators. The use of an animal model allows the study of oocyte maturation failure independent of covariates and confounders, such as pre-existing conditions of the female, which is a significant concern in human studies. Depending on the legislation, it may not be possible to collect and study oocytes from healthy women; and using surplus oocytes from patients undergoing ART may introduce confounders that vary from case to case. FTM oocytes were at various stages of meiotic progression, so correlates of specific times of arrest are not revealed. All the FTM oocytes failed to respond appropriately to an ovulatory stimulus in vivo. Therefore, this analysis informs us about common transcriptome features associated with meiotic incompetence.
Wider implications of the findings: These results reveal that some diagnostic markers of oocyte quality may not reflect developmental competence because even meiotically incompetent oocytes display many normal gene expression features. The results also reveal potential mechanisms by which maternal and environmental factors may impact transcriptional repression and cytoplasmic maturation, and prevent oocyte maturation.
Study funding/competing interest(s): This work was supported by grants from the National Institutes of Health Office of Research Infrastructure Programs Division of Comparative Medicine Grants R24 [OD012221 to K.E.L., OD011107/RR00169 (California National Primate Research Center), and OD010967/RR025880 to C.A.V.]; the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under the award number T32HD087166; MSU AgBioResearch, Michigan State University. Authors have nothing to disclose.
Figures


Similar articles
-
Differing molecular response of young and advanced maternal age human oocytes to IVM.Hum Reprod. 2017 Nov 1;32(11):2199-2208. doi: 10.1093/humrep/dex284. Hum Reprod. 2017. PMID: 29025019 Free PMC article.
-
Human cumulus-enclosed germinal vesicle oocytes from early antral follicles reveal heterogeneous cellular and molecular features associated with in vitro maturation capacity.Hum Reprod. 2015 Jun;30(6):1396-409. doi: 10.1093/humrep/dev083. Epub 2015 Apr 22. Hum Reprod. 2015. PMID: 25904637
-
Endothelin-1 promotes human germinal vesicle-stage oocyte maturation by downregulating connexin-26 expression in cumulus cells.Mol Hum Reprod. 2018 Jan 1;24(1):27-36. doi: 10.1093/molehr/gax058. Mol Hum Reprod. 2018. PMID: 29126233
-
Transcriptomic analysis and epigenetic regulators in human oocytes at different stages of oocyte meiotic maturation.Dev Biol. 2025 Mar;519:55-64. doi: 10.1016/j.ydbio.2024.12.004. Epub 2024 Dec 15. Dev Biol. 2025. PMID: 39681207 Review.
-
Transcriptomic profiles reveal the characteristics of oocytes and cumulus cells at GV, MI, and MII in follicles before ovulation.J Ovarian Res. 2023 Nov 22;16(1):225. doi: 10.1186/s13048-023-01291-2. J Ovarian Res. 2023. PMID: 37993893 Free PMC article. Review.
Cited by
-
Aberrant spliceosome expression and altered alternative splicing events correlate with maturation deficiency in human oocytes.Cell Cycle. 2020 Sep;19(17):2182-2194. doi: 10.1080/15384101.2020.1799295. Epub 2020 Aug 11. Cell Cycle. 2020. PMID: 32779509 Free PMC article.
-
Effects of early lactation body condition loss in dairy cows on serum lipid profiles and on oocyte and cumulus cell transcriptomes.J Dairy Sci. 2022 Oct;105(10):8470-8484. doi: 10.3168/jds.2022-21919. Epub 2022 Aug 6. J Dairy Sci. 2022. PMID: 35940920 Free PMC article.
-
Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans.Nat Commun. 2020 Oct 1;11(1):4917. doi: 10.1038/s41467-020-18680-6. Nat Commun. 2020. PMID: 33004802 Free PMC article.
-
Shared aspects of mRNA expression associated with oocyte maturation failure in humans and rhesus monkeys indicating compromised oocyte quality.Physiol Genomics. 2021 Apr 1;53(4):137-149. doi: 10.1152/physiolgenomics.00155.2020. Epub 2021 Feb 8. Physiol Genomics. 2021. PMID: 33554756 Free PMC article.
-
Follicular Hyperstimulation Dysgenesis: New Explanation for Adverse Effects of Excessive FSH in Ovarian Stimulation.Endocrinology. 2022 Sep 1;163(9):bqac100. doi: 10.1210/endocr/bqac100. Endocrinology. 2022. PMID: 35833461 Free PMC article.
References
-
- Chaffin CL, Vandevoort CA. Follicle growth, ovulation, and luteal formation in primates and rodents: a comparative perspective. Exp Biol Med (Maywood) 2013;238:539–548. - PubMed
-
- Chandra A, Copen CE, Stephen EH. Infertility and impaired fecundity in the United States, 1982–2010: data from the National Survey of Family Growth. Natl Health Stat Report 2013;67:1–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

