Effects of host genetics and environmental conditions on fecal microbiota composition of pigs
- PMID: 30086169
- PMCID: PMC6080793
- DOI: 10.1371/journal.pone.0201901
Effects of host genetics and environmental conditions on fecal microbiota composition of pigs
Abstract
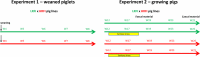
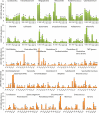
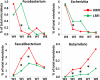
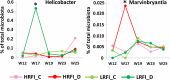
Since microbiota may influence the physiology of its host including body weight increase, growth rate or feed intake, in this study we determined the microbiota composition in high or low residual feed intake (HRFI and LRFI) pig lines, of different age and/or subjected to sanitary stress by sequencing the V3/V4 variable region of 16S rRNA genes. Allisonella, Megasphaera, Mitsuokella, Acidaminococcus (all belonging to Firmicutes/class Negativicutes), Lactobacillus, Faecalibacterium, Catenibacterium, Butyrivibrio, Erysipelotrichaceae, Holdemania, Olsenella and Collinsella were more abundant in HRFI pigs. On the other hand, 26 genera including Bacteroides, Clostridium sensu stricto, Oscillibacter, Paludibacter, Elusimicrobium, Bilophila, Pyramidobacter and TM7 genera, and Clostridium XI and Clostridium XIVa clusters were more abundant in LRFI than HRFI pigs. Adaptation of microbiota to new diet after weaning was slower in LRFI than in HRFI pigs. Sanitary stress was of relatively minor influence on pig microbiota composition in both tested lines although abundance of Helicobacter increased in LRFI pigs subjected to stress. Selection for residual feed intake thus resulted in a selection of fecal microbiota of different composition. However, we cannot conclude whether residual feed intake was directly affected by different microbiota composition or whether the residual feed intake and microbiota composition are two independent consequences of yet unknown genetic traits differentially selected in the pigs of the two lines.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

