CTCF maintains regulatory homeostasis of cancer pathways
- PMID: 30086769
- PMCID: PMC6081938
- DOI: 10.1186/s13059-018-1484-3
CTCF maintains regulatory homeostasis of cancer pathways
Abstract
Background: CTCF binding to DNA helps partition the mammalian genome into discrete structural and regulatory domains. Complete removal of CTCF from mammalian cells causes catastrophic genome dysregulation, likely due to widespread collapse of 3D chromatin looping and alterations to inter- and intra-TAD interactions within the nucleus. In contrast, Ctcf hemizygous mice with lifelong reduction of CTCF expression are viable, albeit with increased cancer incidence. Here, we exploit chronic Ctcf hemizygosity to reveal its homeostatic roles in maintaining genome function and integrity.
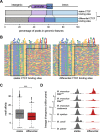
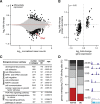
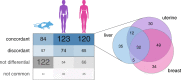
Results: We find that Ctcf hemizygous cells show modest but robust changes in almost a thousand sites of genomic CTCF occupancy; these are enriched for lower affinity binding events with weaker evolutionary conservation across the mouse lineage. Furthermore, we observe dysregulation of the expression of several hundred genes, which are concentrated in cancer-related pathways, and are caused by changes in transcriptional regulation. Chromatin structure is preserved but some loop interactions are destabilized; these are often found around differentially expressed genes and their enhancers. Importantly, the transcriptional alterations identified in vitro are recapitulated in mouse tumors and also in human cancers.
Conclusions: This multi-dimensional genomic and epigenomic profiling of a Ctcf hemizygous mouse model system shows that chronic depletion of CTCF dysregulates steady-state gene expression by subtly altering transcriptional regulation, changes which can also be observed in primary tumors.
Keywords: CTCF; Cancer; Chromatin architecture; Chromatin state; Hemizygosity; Transcription.
Conflict of interest statement
Ethics approval
All animal procedures were conducted in accordance with project (70/7535) and personal licenses, revised by the Animal Welfare and Ethical Review Body at Cancer Research UK Cambridge Institute and issued under the United Kingdom Animals (Scientific Procedures) Act, 1986.
Consent for publication
Not applicable.
Competing interests
PF is a member of the Scientific Advisory Boards of Fabric Genomics, Inc., and Eagle Genomics, Ltd. All other authors have no competing interests to declare.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, et al. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13(12):7612–7624. doi: 10.1128/MCB.13.12.7612. - DOI - PMC - PubMed
-
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16(6):2802–2813. doi: 10.1128/MCB.16.6.2802. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

