One-step CRISPR/Cas9 method for the rapid generation of human antibody heavy chain knock-in mice
- PMID: 30087111
- PMCID: PMC6138433
- DOI: 10.15252/embj.201899243
One-step CRISPR/Cas9 method for the rapid generation of human antibody heavy chain knock-in mice
Abstract
Here, we describe a one-step, in vivo CRISPR/Cas9 nuclease-mediated strategy to generate knock-in mice. We produced knock-in (KI) mice wherein a 1.9-kb DNA fragment bearing a pre-arranged human B-cell receptor heavy chain was recombined into the native murine immunoglobulin locus. Our methodology relies on Cas9 nuclease-induced double-stranded breaks directed by two sgRNAs to occur within the specific target locus of fertilized oocytes. These double-stranded breaks are subsequently repaired via homology-directed repair by a plasmid-borne template containing the pre-arranged human immunoglobulin heavy chain. To validate our knock-in mouse model, we examined the expression of the KI immunoglobulin heavy chains by following B-cell development and performing single B-cell receptor sequencing. We optimized this strategy to generate immunoglobulin KI mice in a short amount of time with a high frequency of homologous recombination (30-50%). In the future, we envision that such knock-in mice will provide much needed vaccination models to evaluate immunoresponses against immunogens specific for various infectious diseases.
Keywords: CRISPR; B cells; antibody responses; bnAbs; knock‐in.
© 2018 The Authors.
Figures

Schematic depicting CRISPR/Cas9 injection. A circular plasmid bearing germline PGT121‐gH VDJ sequences, two guide RNAs, and Cas9 protein were injected into zygotes and implanted into pseudopregnant mice. Cas9‐induced double‐stranded breaks in the genome of zygotes are used to insert germline PGT121 VDJ sequences flanked by homologous arms on each side of the cut site via HDR. After 3 weeks, F0 founder mice are born, some of which bear the human bnAbs germline precursor.
Strategy for insertion of PGT121 rearranged VDJ into mouse IgH locus. Targeting DNA donor with 5′ (3.9 kb) and 3′ (2.6 kb) homology arms to the C57BL/6 WT mouse IgH locus, murine VHJ558 promoter, leader, and the human PGT121 heavy chain VDJ sequences are located between two homology arms. CRISPR/Cas9‐mediated HDR leads to the insertion of the promoter and PGT121 sequences into the C57BL/6 mouse genome. P: murine VHJ558 promoter; HDR: homology‐directed repair; bnAbs: broadly neutralizing antibodies.
sgRNA targeting sites are indicated in red. Three distinct fragments of genomic DNA were amplified by PCR, and in vitro digestion assay was performed with each of the sgRNAs to validate the efficiency of Cas9‐mediated cleavage.
Analysis sgRNA off‐target effects in unrelated genes. Amplicons corresponding to Aakt, Map3K10, and Nop9 were generated by PCR by using gene‐specific primers. In vitro digestion assay was performed to measure the Cas9‐directed cleavage efficiency.


Schematic of the TaqMan probes and their targeting sites within the WT IgH and PGT121 IgH. T: TaqMan probe.
Schematic showing the annealing sites of primers used to validate PGT121 KI animals. Fo.1F and Fo.2F primers were targeted at promoter region and PGT121 region, respectively, and combined with Re.1R primer targeted to the genomic region after homologous 3′ Arm. KI alleles are predicted to result in the amplification of a Fo.1 fragment (3.3 kb) and Fo.2 fragment (2.8 kb). Genomic DNA was extracted from the F0 founders born after CRISPR injection or from a C57BL/6 (WT) mouse. Long‐range PCR was performed to detect the insertion of the PGT121 VDJ sequences at the correct genomic locus.
Table showing the frequency of the different genotypes of mice generated after CRISPR injection with plasmid donors containing long or short homology arms. # of HDR occurrence indicates the integration of the PGT121 heavy chain in the mouse IgH locus. # of Cas9‐mediated D4‐J4 deletions indicates the efficiency of our sgRNA‐directed Cas9 double‐stranded breaks. HC: heavy chain.

3 TaqMan probes, Ighm‐1 WT, HuIghV‐4 Tg, and KI‐P designed for genotyping.
Schematic showing nomenclatures of WT and PGT121 KI mice according to genotyping results.


Bone marrow cells from WT and PGT121+/WT mice were analyzed by flow cytometry using the gating strategy shown on the left. B‐cell progenitors (B220+) were divided into immature (CD43+) and mature (CD43−) cells on the basis of CD43 expression. Data quantified in the panels on the right show the percentage of live cells in the indicated gates (WT: n = 9, PGT121+/WT: n = 8, mean ± SEM).
Early (CD43+) B‐cell progenitors were subdivided according to CD24 and BP‐1 expression into Hardy populations A (CD24−BP‐1−), B (CD24+BP‐1−), and C (CD24+BP‐1+). Data quantified in the panels on the right show the percentage of CD43+ cells in the indicated gates (WT: n = 6, PGT121+/WT: n = 4, mean ± SEM).
Late (CD43−) B‐cell progenitors were subdivided according to IgM and IgD expression into Hardy populations D (IgM−IgD−), E (IgM+IgDint), and F (IgM+IgD+). The data quantified in the panels on the right show the percentage of CD43− cells in the indicated gates (WT: n = 6, PGT121+/WT: n = 4, mean ± SEM).
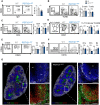
- A
B cells from blood of WT and PGT121+/WT mice were analyzed by flow cytometry. Identification and quantification of B‐cell population (B220+IgD+). Data quantified in the panels on the right show the percentage of live cells in the indicated gates (WT: n = 3, PGT121+/WT: n = 14, mean ± SEM).
- B–E
Spleens from WT and PGT121+/WT mice were analyzed by flow cytometry. (B, C) Identification and quantification of B cells (B220+TCRβ−) and T cells (B220−TCRβ+). T cells were subdivided into CD4 (CD4+CD8−) and CD8 (CD4−CD8+) T cells. Quantification—right. (D, E) B cells were divided on the basis of CD21, CD23, and CD24 expression into T0/T1 cells (CD21−CD24hi), follicular B cells (CD21loCD24lo), T2 cells (CD21hiCD24hiCD23−), marginal zone B (MZB) cells (CD21hiCD24hiCD23+), and quantified (right panels) (n = 5/group, mean ± SEM).
- F
Peritoneal lavage was performed on WT and PGT121+/WT mice, and the exudate was analyzed by flow cytometry. B cells (IgM+) were divided into B2 (CD11b−CD5−), B1b (CD11b+CD5−), and B1a (CD11b+CD5+). Data quantified in the panels on the right show the percentage of IgM+ cells in the indicated gates (WT: n = 8, PGT121+/WT: n = 4, mean ± SEM).
- G
Confocal images spleen cryosections showing the organization of the spleen and the localization of T cells, macrophages, and B cells. Spleen was collected from WT and PGT121+/WT mice. Sections were stained with B220 (blue, B cells), CD169 (red, metallophilic macrophages), TCRβ (green, T cells), and F4/80 (white, red pulp macrophages). In the insets, sequential sections were stained for B220 (blue), CD169 (white), IgM (green), and CD23 (red) to identify follicular (Fo, IgMlowCD23high) and marginal zone (MZ, IgMhighCD23−) B cells. TC: T cells, BC: B cells, Fo: follicular B cells, MZ: marginal zone.

Bone marrow cells from PGT121+/WT mice were divided into late B cell (B220intCD43low) and early B cells (B220int/hiCD43+) on the basis of CD43 expression. B cells from blood of PGT121+/WT mice were stained for IgD and B220, and spleens from PGT121+/WT KI mice stained for B220 and IgM. Ig heavy chains from B cells obtained after single‐cell sorting were PCR amplified and sequenced. The resulting IGHV libraries from bone marrow, blood, and spleen were compared to our PGT121 sequence to determine sequence identity. The pie charts indicate the frequency of IGHV sequences identical to human PGT121 (blue) and murine IGHV (gray).
IgH sequences from early immature, bone marrow (B220+CD43+) B cells. Sequence alignments of 3× representative sequences matching PGT121 from each mouse. Dashes represent identity to the germline reference sequence. FR, framework region; CDR, complementarity‐determining region. Amino acid position is indicated with numbers on top of each reference sequence.

- A, B
Bone marrow B cells from two PGT121+/WT mice were stained for B220 and CD43. Late B cells (B220intCD43low) were single‐cell sorted for antibody heavy chain and subjected to PCR amplification and sequencing. 2/7 (or 28.5%) and 2/9 (22.2%) of the murine IGHV sequences isolated from late B cells exhibited CDR3 motif of PGT121, depicted in blue lettering.
- C
The four murine IGHV sequences were aligned to the germline‐reverted PGT121 heavy chain (blue). The CDR3 motif HGITIFGVVAFKEYYYYYYMDVW of PGT121 was found in all four murine IGHV sequences (indicated by blue arrow). CDR: complementarity‐determining region, FR: framework region.
- D
B cells from peripheral blood of WT or PGT121+/− mice were analyzed by flow cytometry. Identification and quantification of B‐cell population (B220+IgD+) (WT: n = 3, PGT121+/−: n = 2, mean ± SEM)
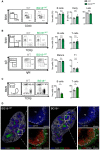
Bone marrow cells from WT and BG18+/WT mice were analyzed by flow cytometry using the gating strategy shown on the left. B‐cell progenitors (B220+) were divided into immature (CD43+) and mature (CD43−) cells on the basis of CD43 expression. Data quantified in the panels on the right show the percentage of live cells in the indicated gates (n = 3/group, mean ± SEM)
Blood from WT and BG18+/WT mice was analyzed by flow cytometry. Identification and quantification of B cells (B220+TCRβ−) and T cells (B220−TCRβ+). B cells were subdivided into mature (IgD+) and T1 (IgD−IgM+) B cells. Quantification—right (WT: n = 4, BG18+/WT: n = 9, mean ± SEM)
B cells from blood of WT or BG18+/− mice were analyzed by flow cytometry. Identification and quantification of B cells (B220+TCRβ−) and T cells (B220−TCRβ+) (WT: n = 6, BG18+/WT: n = 3, mean ± SEM).
Confocal images spleen cryosections showing the organization of the spleen and the localization of T cells, macrophages, and B cells. Spleen was collected from BG18+/WT and BG18+/− mice. Sections were stained with B220 (blue, B cells), CD169 (red, metallophilic macrophages), TCRβ (green, T cells), and F4/80 (white, red pulp macrophages). In the insets, sequential sections were stained for B220 (blue), CD169 (white), IgM (green), and CD23 (red) to identify follicular (Fo, IgMlowCD23high) and marginal zone (MZ, IgMhighCD23−) B cells. TC: T cells, BC: B cells, Fo: follicular B cells, MZ: marginal zone.

B cells from spleens from BG18+/WT or BG18+/− KI mice stained for B220 and IgM. Ig heavy chains from B cells obtained after single‐cell sorting were PCR amplified and sequenced. The resulting IGHV libraries from spleen were compared to our BG18 sequence to determine sequence identity. The pie charts indicate the frequency of IGHV sequences identical to human BG18 (green) and murine IGHV (gray).
IgH sequences from splenic (B220+IgM+) B cells. Sequence alignments of 3× representative sequences matching BG18 from each mouse. Dashes represent identity to the germline reference sequence. FR, framework region; CDR, complementarity‐determining region. Amino acid position is indicated with numbers on top of each reference sequence.

Splenic B cells from three BG18 mice were stained for B220 and IgM. B cells were single‐cell sorted for antibody heavy chain and subjected to PCR amplification and sequencing. The murine IGHV sequences isolated from splenic B cells exhibited CDR3 motif of BG18, depicted in green lettering.
The eight murine IGHV sequences (from Fig 8A; BG18+/−) were aligned to the germline‐reverted BG18 heavy chain (green). The CDR3 motif GVVALGEYYYYGMDVW of BG18 was found in all of these IGHV sequences (indicated by green arrow). CDR: complementarity‐determining region, FR: framework region.
References
-
- Aman MJ, Ravichandran KS (2000) A requirement for lipid rafts in B cell receptor induced Ca(2+) flux. Curr Biol 10: 393–396 - PubMed
-
- Awatramani R, Soriano P, Mai JJ, Dymecki S (2001) An Flp indicator mouse expressing alkaline phosphatase from the ROSA26 locus. Nat Genet 29: 257–259 - PubMed
-
- Benschop RJ, Aviszus K, Zhang X, Manser T, Cambier JC, Wysocki LJ (2001) Activation and anergy in bone marrow B cells of a novel immunoglobulin transgenic mouse that is both hapten specific and autoreactive. Immunity 14: 33–43 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

