Adaptive mutation F772S-enhanced p7-NS4A cooperation facilitates the assembly and release of hepatitis C virus and is associated with lipid droplet enlargement
- PMID: 30087320
- PMCID: PMC6081454
- DOI: 10.1038/s41426-018-0140-z
Adaptive mutation F772S-enhanced p7-NS4A cooperation facilitates the assembly and release of hepatitis C virus and is associated with lipid droplet enlargement
Abstract
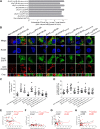
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis and liver cancer worldwide. Adaptive mutations play important roles in the development of the HCV replicon and its infectious clones. We and others have previously identified the p7 mutation F772S and the co-presence of NS4A mutations in infectious HCV full-length clones and chimeric recombinants. However, the underlying mechanism of F772S function remains incompletely understood. Here, we investigated the functional role of F772S using an efficient JFH1-based reporter virus with Core-NS2 from genotype 2a strain J6, and we designated J6-p7/JFH1-4A according to the strain origin of the p7 and NS4A sequences. We found that replacing JFH1-4A with J6-4A (wild-type or mutated NS4A) or genotype 2b J8-4A severely attenuated the viability of J6-p7/JFH1-4A. However, passage-recovered viruses that contained J6-p7 all acquired F772S. Introduction of F772S efficiently rescued the viral spread and infectivity titers of J6-p7/J6-4A, which reached the levels of the original J6-p7/JFH1-4A and led to a concomitant increase in RNA replication, assembly and release of viruses with J6-specific p7 and NS4A. These data suggest that an isolate-specific cooperation existed between p7 and NS4A. NS4A exchange- or substitution-mediated viral attenuation was attributed to the RNA sequence, and no p7-NS4A protein interaction was detected. Moreover, we found that F772S-enhanced p7-NS4A cooperation was associated with the enlargement of intracellular lipid droplets. This study therefore provides new insights into the mechanisms of adaptive mutations and facilitates studies on the HCV life cycle and virus-host interaction.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- The World Health Organization. Global hepatitis report, 2017, <http://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/> (2017).
-
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
