Functional Annotation of ESR1 Gene Fusions in Estrogen Receptor-Positive Breast Cancer
- PMID: 30089255
- PMCID: PMC6171747
- DOI: 10.1016/j.celrep.2018.07.009
Functional Annotation of ESR1 Gene Fusions in Estrogen Receptor-Positive Breast Cancer
Abstract
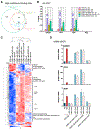
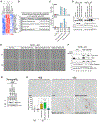
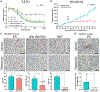
RNA sequencing (RNA-seq) detects estrogen receptor alpha gene (ESR1) fusion transcripts in estrogen receptor-positive (ER+) breast cancer, but their role in disease pathogenesis remains unclear. We examined multiple ESR1 fusions and found that two, both identified in advanced endocrine treatment-resistant disease, encoded stable and functional fusion proteins. In both examples, ESR1-e6>YAP1 and ESR1-e6>PCDH11X, ESR1 exons 1-6 were fused in frame to C-terminal sequences from the partner gene. Functional properties include estrogen-independent growth, constitutive expression of ER target genes, and anti-estrogen resistance. Both fusions activate a metastasis-associated transcriptional program, induce cellular motility, and promote the development of lung metastasis. ESR1-e6>YAP1- and ESR1-e6>PCDH11X-induced growth remained sensitive to a CDK4/6 inhibitor, and a patient-derived xenograft (PDX) naturally expressing the ESR1-e6>YAP1 fusion was also responsive. Transcriptionally active ESR1 fusions therefore trigger both endocrine therapy resistance and metastatic progression, explaining the association with fatal disease progression, although CDK4/6 inhibitor treatment is predicted to be effective.
Keywords: EMT; ESR1 fusions; PDX; breast cancer; endocrine therapy resistance; metastasis.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Bailey TL, and Elkan C (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol 2, 28–36. - PubMed
-
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. (2006). Genome-wide analysis of estrogen receptor binding sites. Nat. Genet 38, 1289–1297. - PubMed
-
- Ellis MJ, Suman VJ, Hoog J, Lin L, Snider J, Prat A, Parker JS, Luo J, DeSchryver K, Allred DC, et al. (2011). Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype—ACOSOG Z1031. J. Clin. Oncol 29, 2342–2349. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

