Substrate Binding by the Second Sialic Acid-Binding Site of Influenza A Virus N1 Neuraminidase Contributes to Enzymatic Activity
- PMID: 30089692
- PMCID: PMC6158415
- DOI: 10.1128/JVI.01243-18
Substrate Binding by the Second Sialic Acid-Binding Site of Influenza A Virus N1 Neuraminidase Contributes to Enzymatic Activity
Abstract
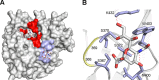
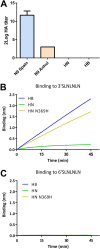
The influenza A virus (IAV) neuraminidase (NA) protein plays an essential role in the release of virus particles from cells and decoy receptors. The NA enzymatic activity presumably needs to match the activity of the IAV hemagglutinin (HA) attachment protein and the host sialic acid (SIA) receptor repertoire. We analyzed the enzymatic activities of N1 NA proteins derived from avian (H5N1) and human (H1N1) IAVs and analyzed the role of the second SIA-binding site, located adjacent to the conserved catalytic site, therein. SIA contact residues in the second SIA-binding site of NA are highly conserved in avian, but not human, IAVs. All N1 proteins preferred cleaving α2,3- over α2,6-linked SIAs even when their corresponding HA proteins displayed a strict preference for α2,6-linked SIAs, indicating that the specificity of the NA protein does not need to fully match that of the corresponding HA protein. NA activity was affected by substitutions in the second SIA-binding site that are observed in avian and human IAVs, at least when multivalent rather than monovalent substrates were used. These mutations included both SIA contact residues and residues that do not directly interact with SIA in all three loops of the second SIA-binding site. Substrate binding via the second SIA-binding site enhanced the catalytic activity of N1. Mutation of the second SIA-binding site was also shown to affect virus replication in vitro Our results indicate an important role for the N1 second SIA-binding site in binding to and cleavage of multivalent substrates.IMPORTANCE Avian and human influenza A viruses (IAVs) preferentially bind α2,3- and α2,6-linked sialic acids (SIAs), respectively. A functional balance between the hemagglutinin (HA) attachment and neuraminidase (NA) proteins is thought to be important for host tropism. What this balance entails at the molecular level is, however, not well understood. We now show that N1 proteins of both avian and human viruses prefer cleaving avian- over human-type receptors although human viruses were relatively better in cleavage of the human-type receptors. In addition, we show that substitutions at different positions in the second SIA-binding site found in NA proteins of human IAVs have a profound effect on binding and cleavage of multivalent, but not monovalent, receptors and affect virus replication. Our results indicate that the HA-NA balance can be tuned via modification of substrate binding via this site and suggest an important role of the second SIA-binding site in host tropism.
Keywords: influenza A virus; neuraminidase; second SIA-binding site; sialic acid.
Copyright © 2018 American Society for Microbiology.
Figures









References
-
- Yen HL, Herlocher LM, Hoffmann E, Matrosovich MN, Monto AS, Webster RG, Govorkova EA. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob Agents Chemother 49:4075–4084. doi:10.1128/AAC.49.10.4075-4084.2005. - DOI - PMC - PubMed
-
- Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. 2015. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 6:e02556-14. doi:10.1128/mBio.02556-14. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

