Quantifying size and diversity of the human T cell alloresponse
- PMID: 30089728
- PMCID: PMC6129121
- DOI: 10.1172/jci.insight.121256
Quantifying size and diversity of the human T cell alloresponse
Abstract
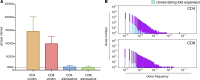
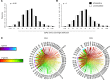
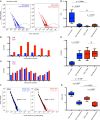
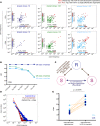
Alloreactive T lymphocytes are the primary mediators of immune responses in transplantation, both in the graft-versus-host and host-versus-graft directions. While essentially all clones comprising the human T cell repertoire have been selected on self-peptide presented by self-human leukocyte antigens (self-HLAs), much remains to be understood about the nature of clones capable of responding to allo-HLA molecules. Quantitative tools to study these cells are critical to understand fundamental features of this important response; however, the large size and diversity of the alloreactive T cell repertoire in humans presents a great technical challenge. We have developed a high-throughput T cell receptor (TCR) sequencing approach to characterize the human alloresponse. We present a statistical method to model T cell clonal frequency distribution and quantify repertoire diversity. Using these approaches, we measured the diversity and frequency of distinct alloreactive CD4+ and CD8+ T cell populations in HLA-mismatched responder-stimulator pairs. Our findings indicate that the alloimmune repertoire is highly specific for a given pair of individuals, that most alloreactive clones circulate at low frequencies, and that a high proportion of TCRs is likely able to recognize alloantigens.
Keywords: Bioinformatics; Immunology; T cells; T-cell receptor; Transplantation.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

