Forcing the reversibility of a mechanochemical reaction
- PMID: 30089863
- PMCID: PMC6082871
- DOI: 10.1038/s41467-018-05115-6
Forcing the reversibility of a mechanochemical reaction
Abstract
Mechanical force modifies the free-energy surface of chemical reactions, often enabling thermodynamically unfavoured reaction pathways. Most of our molecular understanding of force-induced reactivity is restricted to the irreversible homolytic scission of covalent bonds and ring-opening in polymer mechanophores. Whether mechanical force can by-pass thermodynamically locked reactivity in heterolytic bimolecular reactions and how this impacts the reaction reversibility remains poorly understood. Using single-molecule force-clamp spectroscopy, here we show that mechanical force promotes the thermodynamically disfavored SN2 cleavage of an individual protein disulfide bond by poor nucleophilic organic thiols. Upon force removal, the transition from the resulting high-energy unstable mixed disulfide product back to the initial, low-energy disulfide bond reactant becomes suddenly spontaneous, rendering the reaction fully reversible. By rationally varying the nucleophilicity of a series of small thiols, we demonstrate how force-regulated chemical kinetics can be finely coupled with thermodynamics to predict and modulate the reversibility of bimolecular mechanochemical reactions.
Conflict of interest statement
The authors declare no competing interests.
Figures
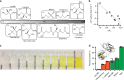
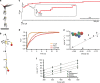
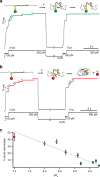
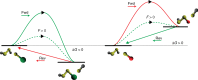
References
-
- Garcia-Manyes S, Beedle AEM. Steering chemical reactions with force. Nat. Rev. Chem. 2017;1:1–16. doi: 10.1038/s41570-017-0083. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

