Predicting Antigen Presentation-What Could We Learn From a Million Peptides?
- PMID: 30090105
- PMCID: PMC6068240
- DOI: 10.3389/fimmu.2018.01716
Predicting Antigen Presentation-What Could We Learn From a Million Peptides?
Abstract
Antigen presentation lies at the heart of immune recognition of infected or malignant cells. For this reason, important efforts have been made to predict which peptides are more likely to bind and be presented by the human leukocyte antigen (HLA) complex at the surface of cells. These predictions have become even more important with the advent of next-generation sequencing technologies that enable researchers and clinicians to rapidly determine the sequences of pathogens (and their multiple variants) or identify non-synonymous genetic alterations in cancer cells. Here, we review recent advances in predicting HLA binding and antigen presentation in human cells. We argue that the very large amount of high-quality mass spectrometry data of eluted (mainly self) HLA ligands generated in the last few years provides unprecedented opportunities to improve our ability to predict antigen presentation and learn new properties of HLA molecules, as demonstrated in many recent studies of naturally presented HLA-I ligands. Although major challenges still lie on the road toward the ultimate goal of predicting immunogenicity, these experimental and computational developments will facilitate screening of putative epitopes, which may eventually help decipher the rules governing T cell recognition.
Keywords: T cell epitope; antigen presentation; computational immunology; human leukocyte antigen ligand prediction; human leukocyte antigen peptidomics.
Figures
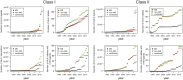
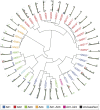
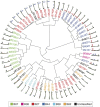

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

