Regulation of Ferredoxin-NADP+ Oxidoreductase to Cyclic Electron Transport in High Salinity Stressed Pyropia yezoensis
- PMID: 30090109
- PMCID: PMC6068275
- DOI: 10.3389/fpls.2018.01092
Regulation of Ferredoxin-NADP+ Oxidoreductase to Cyclic Electron Transport in High Salinity Stressed Pyropia yezoensis
Abstract
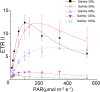
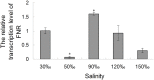
Pyropia yezoensis can survive the severe water loss that occurs during low tide, making it an ideal species to investigate the acclimation mechanism of intertidal seaweed to special extreme environments. In this study, we determined the effects of high salinity on photosynthesis using increasing salinity around algal tissues. Both electron transport rates, ETR (I) and ETR (II), showed continuous decreases as the salinity increased. However, the difference between these factors remained relatively stable, similar to the control. Inhibitor experiments illustrated that there were at least three different cyclic electron transport pathways. Under conditions of severe salinity, NAD(P)H could be exploited as an endogenous electron donor to reduce the plastoquinone pool in Py. yezoensis. Based on these findings, we next examined how these different cyclic electron transport (CETs) pathways were coordinated by cloning the gene (HM370553) for ferredoxin-NADP+ oxidoreductase (FNR). A phylogenetic tree was constructed, and the evolutionary relationships among different FNRs were evaluated. The results indicated that the Py. yezoensis FNR showed a closer relationship with cyanobacterial FNR. The results of both real-time polymerase chain reaction and western blotting showed that the enzyme was upregulated under 90-120‰ salinity. Due to the structure-function correlations in organism, Py. yezoensis FNR was proposed to be involved in NAD(P)H-dependent Fd+ reduction under severe salinity conditions. Thus, through the connection between different donors bridged by FNR, electrons were channeled toward distinct routes according to the different metabolic demands. This was expected to make the electron transfer in the chloroplasts become more flexible and to contribute greatly to acclimation of Py. yezoensis to the extreme variable environments in the intertidal zone.
Keywords: Pyropia yezoensis; electron transportation; environmental acclimation; ferredoxin-NADP+ reductase; stress responding.
Figures







References
-
- Baker N. R. (1991). A possible role for photosystem II in environmental perturbations of photosynthesis. Physiol. Plant. 81 563–570. 10.1111/j.1399-3054.1991.tb05101.x - DOI
-
- Batie C. J., Kamin H. (1984). Ferredoxin-NADP+ oxidoreductase. Equilibria in binary and ternary complexes with NADP+ and ferredoxin. J. Biol. Chem. 259 8832–8839. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

