New enzymatic and mass spectrometric methodology for the selective investigation of gut microbiota-derived metabolites
- PMID: 30090311
- PMCID: PMC6063053
- DOI: 10.1039/c8sc01502c
New enzymatic and mass spectrometric methodology for the selective investigation of gut microbiota-derived metabolites
Abstract
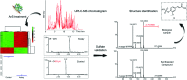
Gut microbiota significantly impact human physiology through metabolic interaction. Selective investigation of the co-metabolism of bacteria and their human host is a challenging task and methods for their analysis are limited. One class of metabolites associated with this co-metabolism are O-sulfated compounds. Herein, we describe the development of a new enzymatic assay for the selective mass spectrometric investigation of this phase II modification class. Analysis of human urine and fecal samples resulted in the detection of 206 sulfated metabolites, which is three times more than reported in the Human Metabolome Database. We confirmed the chemical structure of 36 sulfated metabolites including unknown and commonly reported microbiota-derived sulfated metabolites using synthesized internal standards and mass spectrometric fragmentation experiments. Our findings demonstrate that enzymatic sample pre-treatment combined with state-of-the-art metabolomics analysis represents a new and efficient strategy for the discovery of unknown microbiota-derived metabolites in human samples. Our described approach can be adapted for the targeted investigation of other metabolite classes as well as the discovery of biomarkers for diseases affected by microbiota.
Figures



References
-
- Backhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I. Science. 2005;307:1915–1920. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

