PBPK Models for CYP3A4 and P-gp DDI Prediction: A Modeling Network of Rifampicin, Itraconazole, Clarithromycin, Midazolam, Alfentanil, and Digoxin
- PMID: 30091221
- PMCID: PMC6202474
- DOI: 10.1002/psp4.12343
PBPK Models for CYP3A4 and P-gp DDI Prediction: A Modeling Network of Rifampicin, Itraconazole, Clarithromycin, Midazolam, Alfentanil, and Digoxin
Abstract
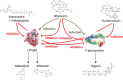
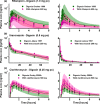
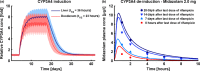
According to current US Food and Drug Administration (FDA) and European Medicines Agency (EMA) guidance documents, physiologically based pharmacokinetic (PBPK) modeling is a powerful tool to explore and quantitatively predict drug-drug interactions (DDIs) and may offer an alternative to dedicated clinical trials. This study provides whole-body PBPK models of rifampicin, itraconazole, clarithromycin, midazolam, alfentanil, and digoxin within the Open Systems Pharmacology (OSP) Suite. All models were built independently, coupled using reported interaction parameters, and mutually evaluated to verify their predictive performance by simulating published clinical DDI studies. In total, 112 studies were used for model development and 57 studies for DDI prediction. 93% of the predicted area under the plasma concentration-time curve (AUC) ratios and 94% of the peak plasma concentration (Cmax ) ratios are within twofold of the observed values. This study lays a cornerstone for the qualification of the OSP platform with regard to reliable PBPK predictions of enzyme-mediated and transporter-mediated DDIs during model-informed drug development. All presented models are provided open-source and transparently documented.
© 2018 The Authors CPT: Pharmacometrics & Systems Pharmacology published by Wiley Periodicals, Inc. on behalf of American Society for Clinical Pharmacology and Therapeutics.
Figures


References
-
- U.S. Food and Drug Administration . Clinical Drug Interaction Studies – Study Design, Data Analysis, and Clinical Implications. Draft Guidance for Industry. <https://www.fda.gov/downloads/drugs/guidances/ucm292362.pdf>, (October 2017).
-
- European Medicines Agency . Guideline on the Investigation of Drug Interactions. CPMP/EWP/560/95/Rev. 1 Corr. 2** <http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guidelin...>, (21 June 2012).
-
- Guengerich, F.P. Cytochrome P‐450 3A4: regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 39, 1–17 (1999). - PubMed
-
- Fenner, K.S. et al Drug‐drug interactions mediated through P‐glycoprotein: clinical relevance and in vitro‐in vivo correlation using digoxin as a probe drug. Clin. Pharmacol. Ther. 85, 173–181 (2009). - PubMed
-
- U.S. Food and Drug Administration . Drug development and drug interactions: table of substrates, inhibitors and inducers. <https://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResource...>.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

