How Acts of Infidelity Promote DNA Break Repair: Collision and Collusion Between DNA Repair and Transcription
- PMID: 30091472
- PMCID: PMC6334755
- DOI: 10.1002/bies.201800045
How Acts of Infidelity Promote DNA Break Repair: Collision and Collusion Between DNA Repair and Transcription
Abstract
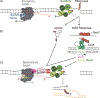
Transcription is a fundamental cellular process and the first step in gene regulation. Although RNA polymerase (RNAP) is highly processive, in growing cells the progression of transcription can be hindered by obstacles on the DNA template, such as damaged DNA. The authors recent findings highlight a trade-off between transcription fidelity and DNA break repair. While a lot of work has focused on the interaction between transcription and nucleotide excision repair, less is known about how transcription influences the repair of DNA breaks. The authors suggest that when the cell experiences stress from DNA breaks, the control of RNAP processivity affects the balance between preserving transcription integrity and DNA repair. Here, how the conflict between transcription and DNA double-strand break (DSB) repair threatens the integrity of both RNA and DNA are discussed. In reviewing this field, the authors speculate on cellular paradigms where this equilibrium is well sustained, and instances where the maintenance of transcription fidelity is favored over genome stability.
Keywords: DNA break repair; DNA resection; RNA polymerase; RecBCD; genome stability; transcription fidelity.
© 2018 WILEY Periodicals, Inc.
Figures

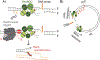
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

