A discriminator code-based DTD surveillance ensures faithful glycine delivery for protein biosynthesis in bacteria
- PMID: 30091703
- PMCID: PMC6097841
- DOI: 10.7554/eLife.38232
A discriminator code-based DTD surveillance ensures faithful glycine delivery for protein biosynthesis in bacteria
Abstract
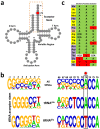
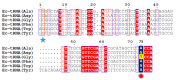
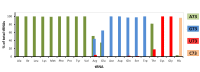
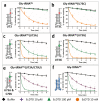
D-aminoacyl-tRNA deacylase (DTD) acts on achiral glycine, in addition to D-amino acids, attached to tRNA. We have recently shown that this activity enables DTD to clear non-cognate Gly-tRNAAla with 1000-fold higher efficiency than its activity on Gly-tRNAGly, indicating tRNA-based modulation of DTD (Pawar et al., 2017). Here, we show that tRNA's discriminator base predominantly accounts for this activity difference and is the key to selection by DTD. Accordingly, the uracil discriminator base, serving as a negative determinant, prevents Gly-tRNAGly misediting by DTD and this protection is augmented by EF-Tu. Intriguingly, eukaryotic DTD has inverted discriminator base specificity and uses only G3•U70 for tRNAGly/Ala discrimination. Moreover, DTD prevents alanine-to-glycine misincorporation in proteins rather than only recycling mischarged tRNAAla. Overall, the study reveals the unique co-evolution of DTD and discriminator base, and suggests DTD's strong selection pressure on bacterial tRNAGlys to retain a pyrimidine discriminator code.
Keywords: E. coli; biochemistry; chemical biology; genetic code; proofreading; protein synthesis; synthetase; tRNA; translation.
© 2018, Kuncha et al.
Conflict of interest statement
SK, KS, KP, JG, SR, SP, SK, RS No competing interests declared
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- DST-INSPIRE/Department of Science and Technology, Ministry of Science and Technology/International
- DBT-RA/Department of Biotechnology , Ministry of Science and Technology/International
- Centre of Excellence/Department of Biotechnology, Ministry of Science and Technology/International
- J. C. Bose Fellowship/Science and Engineering Research Board/International
- Centre of Excellence/Department of Biotechnology , Ministry of Science and Technology/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

