RNA binding protein HuR regulates extracellular matrix gene expression and pH homeostasis independent of controlling HIF-1α signaling in nucleus pulposus cells
- PMID: 30092282
- PMCID: PMC6367062
- DOI: 10.1016/j.matbio.2018.08.003
RNA binding protein HuR regulates extracellular matrix gene expression and pH homeostasis independent of controlling HIF-1α signaling in nucleus pulposus cells
Abstract
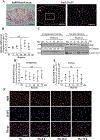
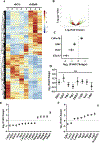
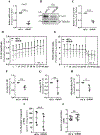
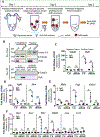
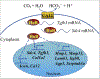
Nucleus pulposus (NP) cells reside in the hypoxic niche of the intervertebral disc. Studies have demonstrated that RNA-binding protein HuR modulates hypoxic signaling in several cancers, however, its function in the disc is unknown. HuR did not show cytoplasmic translocation in hypoxia and its silencing did not alter levels of Hif-1α or HIF-targets in NP cells. RNA-Sequencing data revealed that important extracellular matrix-related genes including several collagens, MMPs, aggrecan, Tgf-β3 and Sdc4 were regulated by HuR. Further analysis of HuR-silenced NP cells confirmed that HuR maintained expression of these matrix genes. We confirmed decreased levels of secreted collagen I and Sdc4 and increased pro-MMP13 in HuR-knockdown cells. In addition, messenger ribonucleoprotein immunoprecipitation demonstrated HuR binding to Tgf-β3 and Sdc4 mRNAs. Interestingly, while HuR bound to Hif-1α and Vegf mRNAs, it was clear that compensatory mechanisms sustained their expression when HuR was silenced. Noteworthy, despite the presence of multiple HuR-binding sites and reported interaction in other cell types, HuR showed no binding to Pgk1, Eno1, Pdk1 and Pfkfb3 in NP cells. Metabolic studies showed a significant decrease in the extracellular acidification rate (ECAR) and mitochondrial oxygen consumption rate (OCR) and acidic pH in HuR-silenced NP cells, without appreciable change in total OCR. These changes were likely due to decreased Ca12 expression in HuR silenced cells. Taken together, our study demonstrates for the first time that HuR regulates extracellular matrix (ECM) and pH homeostasis of NP cells and has important implications in the maintenance of intervertebral disc health.
Keywords: Extracellular matrix (ECM); HIF-1α; Human antigen R (HuR); Intervertebral disc; Nucleus pulposus.
Copyright © 2018 Elsevier B.V. All rights reserved.
Conflict of interest statement
Figures



References
-
- Hoy D, March L, Brooks P, Blyth F, Woolf A, Bain C, Williams G, Smith E, Vos T, Barendregt J, Murray C, Burstein R, Buchbinder R, The global burden of low back pain: estimates from the Global Burden of Disease 2010 study, Ann. Rheum. Dis 73 (2014) 968–974. - PubMed
-
- Balague F, Mannion AF, Pellise F, Cedraschi C, Non-specific low back pain, Lancet 379 (2012) 482–91. - PubMed
-
- Vos T, Barber RM, Bell B, Bertozzi-Villa A, Biryukov S, Bolliger I, Charlson F, Davis A, Degenhardt L, Dicker D, et al., Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013, Lancet 386 (2015) 743–800. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

