Handling heme: The mechanisms underlying the movement of heme within and between cells
- PMID: 30092350
- PMCID: PMC6363905
- DOI: 10.1016/j.freeradbiomed.2018.08.005
Handling heme: The mechanisms underlying the movement of heme within and between cells
Abstract
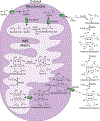
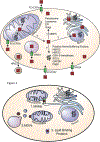
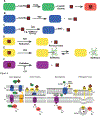
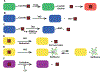
Heme is an essential cofactor and signaling molecule required for virtually all aerobic life. However, excess heme is cytotoxic. Therefore, heme must be safely transported and trafficked from the site of synthesis in the mitochondria or uptake at the cell surface, to hemoproteins in most subcellular compartments. While heme synthesis and degradation are relatively well characterized, little is known about how heme is trafficked and transported throughout the cell. Herein, we review eukaryotic heme transport, trafficking, and mobilization, with a focus on factors that regulate bioavailable heme. We also highlight the role of gasotransmitters and small molecules in heme mobilization and bioavailability, and heme trafficking at the host-pathogen interface.
Keywords: Heme; Heme trafficking; Heme transport; Host-pathogen interactions; Hydrogen peroxide; Iron; Nitric oxide.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure:
The authors declare no conflict of interest.
Figures

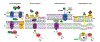
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

