VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites
- PMID: 30093493
- PMCID: PMC6168267
- DOI: 10.1083/jcb.201807019
VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites
Abstract
Mutations in the human VPS13 genes are responsible for neurodevelopmental and neurodegenerative disorders including chorea acanthocytosis (VPS13A) and Parkinson's disease (VPS13C). The mechanisms of these diseases are unknown. Genetic studies in yeast hinted that Vps13 may have a role in lipid exchange between organelles. In this study, we show that the N-terminal portion of VPS13 is tubular, with a hydrophobic cavity that can solubilize and transport glycerolipids between membranes. We also show that human VPS13A and VPS13C bind to the ER, tethering it to mitochondria (VPS13A), to late endosome/lysosomes (VPS13C), and to lipid droplets (both VPS13A and VPS13C). These findings identify VPS13 as a lipid transporter between the ER and other organelles, implicating defects in membrane lipid homeostasis in neurological disorders resulting from their mutations. Sequence and secondary structure similarity between the N-terminal portions of Vps13 and other proteins such as the autophagy protein ATG2 suggest lipid transport roles for these proteins as well.
© 2018 Kumar et al.
Figures
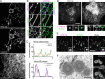
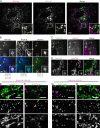

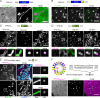

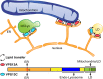
Comment in
-
VPS13: A lipid transfer protein making contacts at multiple cellular locations.J Cell Biol. 2018 Oct 1;217(10):3322-3324. doi: 10.1083/jcb.201808151. Epub 2018 Sep 4. J Cell Biol. 2018. PMID: 30181317 Free PMC article.
References
-
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 - DOI - PMC - PubMed
-
- Alesutan I., Seifert J., Pakladok T., Rheinlaender J., Lebedeva A., Towhid S.T., Stournaras C., Voelkl J., Schäffer T.E., and Lang F.. 2013. Chorein sensitivity of actin polymerization, cell shape and mechanical stiffness of vascular endothelial cells. Cell. Physiol. Biochem. 32:728–742. 10.1159/000354475 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- S10 RR029205/RR/NCRR NIH HHS/United States
- P41 GM103403/GM/NIGMS NIH HHS/United States
- R35 GM131715/GM/NIGMS NIH HHS/United States
- P30 DK045735/DK/NIDDK NIH HHS/United States
- F31 NS110229/NS/NINDS NIH HHS/United States
- T32 GM008616/GM/NIGMS NIH HHS/United States
- R37 NS036251/NS/NINDS NIH HHS/United States
- R01 GM080616/GM/NIGMS NIH HHS/United States
- T32 GM008283/GM/NIGMS NIH HHS/United States
- P30 DA018343/DA/NIDA NIH HHS/United States
- R01 NS036251/NS/NINDS NIH HHS/United States
- T32 GM007205/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

